Diffusion Imaging In Python¶
DIPY is a free and open source software project for computational neuroanatomy, focusing mainly on diffusion magnetic resonance imaging (dMRI) analysis. It implements a broad range of algorithms for denoising, registration, reconstruction, tracking, clustering, visualization, and statistical analysis of MRI data.
Highlights¶
DIPY 1.0.0 is now available. New features include:
Critical API changes
Large refactoring of tracking API.
New denoising algorithm: MP-PCA.
New Gibbs ringing removal.
New interpolation module:
dipy.core.interpolation.New reconstruction models: MTMS-CSD, Mean Signal DKI.
Increased coordinate systems consistency.
New object to manage safely tractography data: StatefulTractogram
New command line interface for downloading datasets: FetchFlow
Horizon updated, medical visualization interface powered by QuickBundlesX.
Removed all deprecated functions and parameters.
Removed compatibility with Python 2.7.
Updated minimum dependencies version (Numpy, Scipy).
All tutorials updated to API changes and 3 new added.
Large documentation update.
Closed 289 issues and merged 98 pull requests.
See Older Highlights.
Announcements¶
DIPY Workshop - Titanium Edition (March 11-15, 2019) is now open for registration:
DIPY 1.0 released August 5, 2019.
DIPY 0.16 released March 10, 2019.
DIPY 0.15 released December 12, 2018.
See some of our Past Announcements
Getting Started¶
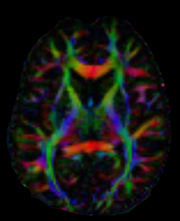
Here is a quick snippet showing how to calculate color FA also known as the DEC map. We use a Tensor model to reconstruct the datasets which are saved in a Nifti file along with the b-values and b-vectors which are saved as text files. Finally, we save our result as a Nifti file
fdwi = 'dwi.nii.gz'
fbval = 'dwi.bval'
fbvec = 'dwi.bvec'
from dipy.io.image import load_nifti, save_nifti
from dipy.io import read_bvals_bvecs
from dipy.core.gradients import gradient_table
from dipy.reconst.dti import TensorModel
data, affine = load_nifti(fdwi)
bvals, bvecs = read_bvals_bvecs(fbval, fbvec)
gtab = gradient_table(bvals, bvecs)
tenmodel = TensorModel(gtab)
tenfit = tenmodel.fit(data)
save_nifti('colorfa.nii.gz', tenfit.color_fa, affine)
As an exercise, you can try to calculate color FA with your datasets. You will need to replace the filepaths fdwi, fbval and fbvec. Here is what a slice should look like.
Next Steps¶
You can learn more about how you to use DIPY with your datasets by reading the examples in our Documentation.
Support¶
We acknowledge support from the following organizations:
The department of Intelligent Systems Engineering of Indiana University.
The National Institute of Biomedical Imaging and Bioengineering, NIH.
The Gordon and Betty Moore Foundation and the Alfred P. Sloan Foundation, through the University of Washington eScience Institute Data Science Environment.
Google supported DIPY through the Google Summer of Code Program during Summer 2015, 2016 and 2018.
The International Neuroinformatics Coordination Facility.