Automatic Fiber Bundle Extraction with RecoBundles¶
This example explains how we can use RecoBundles [Garyfallidis17] to extract bundles from tractograms.
First import the necessary modules.
from dipy.data.fetcher import get_two_hcp842_bundles
from dipy.data.fetcher import (fetch_target_tractogram_hcp,
fetch_bundle_atlas_hcp842,
get_bundle_atlas_hcp842,
get_target_tractogram_hcp)
import numpy as np
from dipy.segment.bundles import RecoBundles
from dipy.align.streamlinear import whole_brain_slr
from fury import actor, window
from dipy.io.stateful_tractogram import Space, StatefulTractogram
from dipy.io.streamline import load_trk, save_trk
from dipy.io.utils import create_tractogram_header
Download and read data for this tutorial
target_file, target_folder = fetch_target_tractogram_hcp()
atlas_file, atlas_folder = fetch_bundle_atlas_hcp842()
atlas_file, all_bundles_files = get_bundle_atlas_hcp842()
target_file = get_target_tractogram_hcp()
sft_atlas = load_trk(atlas_file, "same", bbox_valid_check=False)
atlas = sft_atlas.streamlines
atlas_header = create_tractogram_header(atlas_file,
*sft_atlas.space_attribute)
sft_target = load_trk(target_file, "same", bbox_valid_check=False)
target = sft_target.streamlines
target_header = create_tractogram_header(atlas_file,
*sft_atlas.space_attribute)
let’s visualize atlas tractogram and target tractogram before registration
interactive = False
ren = window.Renderer()
ren.SetBackground(1, 1, 1)
ren.add(actor.line(atlas, colors=(1, 0, 1)))
ren.add(actor.line(target, colors=(1, 1, 0)))
window.record(ren, out_path='tractograms_initial.png', size=(600, 600))
if interactive:
window.show(ren)
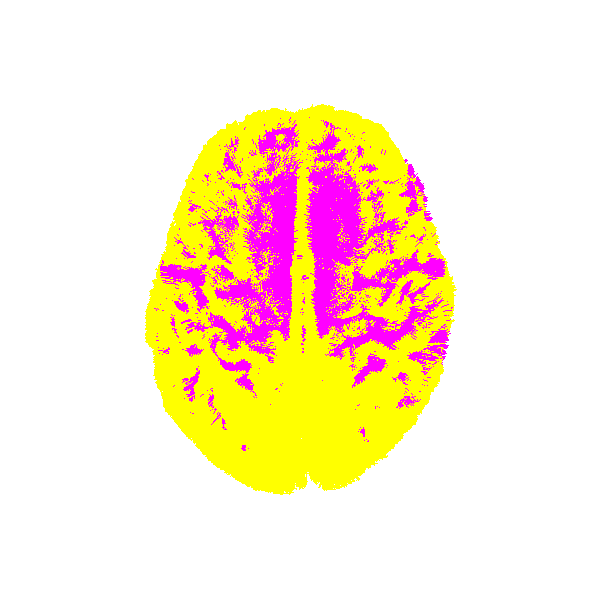
Atlas and target before registration.¶
We will register target tractogram to model atlas’ space using streamlinear registeration (SLR) [Garyfallidis15]
moved, transform, qb_centroids1, qb_centroids2 = whole_brain_slr(
atlas, target, x0='affine', verbose=True, progressive=True)
We save the transform generated in this registration, so that we can use it in the bundle profiles example
np.save("slr_transform.npy", transform)
let’s visualize atlas tractogram and target tractogram after registration
interactive = False
ren = window.Renderer()
ren.SetBackground(1, 1, 1)
ren.add(actor.line(atlas, colors=(1, 0, 1)))
ren.add(actor.line(moved, colors=(1, 1, 0)))
window.record(ren, out_path='tractograms_after_registration.png',
size=(600, 600))
if interactive:
window.show(ren)
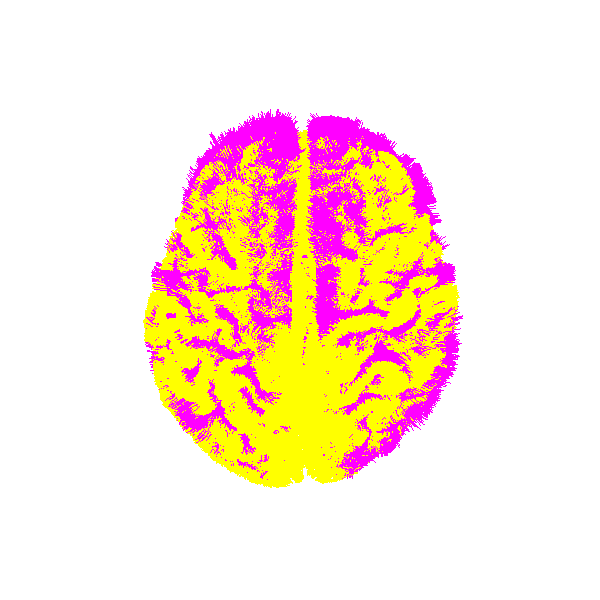
Atlas and target after registration.¶
Read AF left and CST left bundles from already fetched atlas data to use them as model bundles
model_af_l_file, model_cst_l_file = get_two_hcp842_bundles()
Extracting bundles using recobundles [Garyfallidis17]
sft_af_l = load_trk(model_af_l_file, "same", bbox_valid_check=False)
model_af_l = sft_af_l.streamlines
rb = RecoBundles(moved, verbose=True)
recognized_af_l, af_l_labels = rb.recognize(model_bundle=model_af_l,
model_clust_thr=5.,
reduction_thr=10,
reduction_distance='mam',
slr=True,
slr_metric='asymmetric',
pruning_distance='mam')
let’s visualize extracted Arcuate Fasciculus Left bundle and model bundle together
interactive = False
ren = window.Renderer()
ren.SetBackground(1, 1, 1)
ren.add(actor.line(model_af_l, colors=(.1, .7, .26)))
ren.add(actor.line(recognized_af_l, colors=(.1, .1, 6)))
ren.set_camera(focal_point=(320.21296692, 21.28884506, 17.2174015),
position=(2.11, 200.46, 250.44), view_up=(0.1, -1.028, 0.18))
window.record(ren, out_path='AF_L_recognized_bundle.png',
size=(600, 600))
if interactive:
window.show(ren)

Extracted Arcuate Fasciculus Left bundle and model bundle¶
Save the bundle as a trk file. Rather than saving the recognized streamlines in the space of the atlas, we save the streamlines that are in the original space of the subject anatomy.
reco_af_l = StatefulTractogram(target[af_l_labels], target_header,
Space.RASMM)
save_trk(reco_af_l, "AF_L.trk", bbox_valid_check=False)
sft_cst_l = load_trk(model_cst_l_file, "same", bbox_valid_check=False)
model_cst_l = sft_cst_l.streamlines
recognized_cst_l, cst_l_labels = rb.recognize(model_bundle=model_cst_l,
model_clust_thr=5.,
reduction_thr=10,
reduction_distance='mam',
slr=True,
slr_metric='asymmetric',
pruning_distance='mam')
let’s visualize extracted Corticospinal Tract (CST) Left bundle and model bundle together
interactive = False
ren = window.Renderer()
ren.SetBackground(1, 1, 1)
ren.add(actor.line(model_cst_l, colors=(.1, .7, .26)))
ren.add(actor.line(recognized_cst_l, colors=(.1, .1, 6)))
ren.set_camera(focal_point=(-18.17281532, -19.55606842, 6.92485857),
position=(-360.11, -340.46, -40.44),
view_up=(-0.03, 0.028, 0.89))
window.record(ren, out_path='CST_L_recognized_bundle.png',
size=(600, 600))
if interactive:
window.show(ren)

Extracted Corticospinal Tract (CST) Left bundle and model bundle¶
Save the bundle as a trk file:
reco_cst_l = StatefulTractogram(target[cst_l_labels], target_header,
Space.RASMM)
save_trk(reco_cst_l, "CST_L.trk", bbox_valid_check=False)
References¶
- Garyfallidis17(1,2)
Garyfallidis et al. Recognition of white matter bundles using local and global streamline-based registration and clustering, Neuroimage, 2017.
Example source code
You can download the full source code of this example. This same script is also included in the dipy source distribution under the doc/examples/ directory.