Introduction to command line interfaces#
This tutorial provides a basic introduction to DIPY’s [1] command line interfaces.
Using a terminal, let’s download a dataset. This is multi-shell dataset, which was kindly provided by Hansen and Jespersen (more details about the data are provided in their paper [2]). For this tutorial we will use a Linux terminal, please adapt accordingly if you are using Mac or Windows.
First let’s create a folder:
mkdir data_folder
Download the data in the data_folder:
dipy_fetch cfin_multib --out_dir data_folder
Move to the folder with the data:
cd data_folder/cfin_multib
ls
__DTI_AX_ep2d_2_5_iso_33d_20141015095334_4.bval
__DTI_AX_ep2d_2_5_iso_33d_20141015095334_4.bvec
__DTI_AX_ep2d_2_5_iso_33d_20141015095334_4.nii
T1.nii
Let’s rename the long filenames to something that is easier to read:
mv __DTI_AX_ep2d_2_5_iso_33d_20141015095334_4.bval dwi.bval
mv __DTI_AX_ep2d_2_5_iso_33d_20141015095334_4.bvec dwi.bvec
mv __DTI_AX_ep2d_2_5_iso_33d_20141015095334_4.nii dwi.nii
We can use dipy_info to check the bval and nii files
dipy_info *.bval *.nii
INFO:-----------------
INFO:Looking at T1.nii
INFO:-----------------
INFO:Data size (256, 256, 176)
INFO:Data type uint16
INFO:Data min 0 max 888 avg 62.940581408413976
INFO:2nd percentile 0.0 98th percentile 377.0
INFO:Native coordinate system PSR
INFO:Affine Native to RAS matrix
[[ 0. 0.01 1. -89.569]
[ -1. 0. 0. 138.451]
[ 0. 1. -0.01 -131.289]
[ 0. 0. 0. 1. ]]
INFO:Voxel size [1. 1. 1.]
INFO:---------------------------------------------------------
INFO:Looking at __DTI_AX_ep2d_2_5_iso_33d_20141015095334_4.nii
INFO:---------------------------------------------------------
INFO:Data size (96, 96, 19, 496)
INFO:Data type int16
INFO:Data min 0 max 1257 avg 58.62918037280702 of vol 0
INFO:2nd percentile 0.0 98th percentile 234.0 of vol 0
INFO:Native coordinate system LAS
INFO:Affine Native to RAS matrix
[[ -2.498 0.084 0.067 113.641]
[ 0.069 2.451 -0.488 -104.142]
[ 0.082 0.486 2.451 -31.504]
[ 0. 0. 0. 1. ]]
INFO:Voxel size [2.5 2.5 2.5]
(base) C:\Users\elef\Data\examples\data_folder\cfin_multib>dipy_info *.bval *.nii
INFO:----------------------------------------------------------
INFO:Looking at __DTI_AX_ep2d_2_5_iso_33d_20141015095334_4.bval
INFO:----------------------------------------------------------
INFO:b-values
[ 0. 200. 200. 200. 200. 200. 200. 200. 200. 200. 200. 200.
200. 200. 200. 200. 200. 200. 200. 200. 200. 200. 200. 200.
200. 200. 200. 200. 200. 200. 200. 200. 200. 200. 400. 400.
400. 400. 400. 400. 400. 400. 400. 400. 400. 400. 400. 400.
400. 400. 400. 400. 400. 400. 400. 400. 400. 400. 400. 400.
400. 400. 400. 400. 400. 400. 400. 600. 600. 600. 600. 600.
600. 600. 600. 600. 600. 600. 600. 600. 600. 600. 600. 600.
600. 600. 600. 600. 600. 600. 600. 600. 600. 600. 600. 600.
600. 600. 600. 600. 800. 800. 800. 800. 800. 800. 800. 800.
800. 800. 800. 800. 800. 800. 800. 800. 800. 800. 800. 800.
800. 800. 800. 800. 800. 800. 800. 800. 800. 800. 800. 800.
800. 1000. 1000. 1000. 1000. 1000. 1000. 1000. 1000. 1000. 1000. 1000.
1000. 1000. 1000. 1000. 1000. 1000. 1000. 1000. 1000. 1000. 1000. 1000.
1000. 1000. 1000. 1000. 1000. 1000. 1000. 1000. 1000. 1000. 1200. 1200.
1200. 1200. 1200. 1200. 1200. 1200. 1200. 1200. 1200. 1200. 1200. 1200.
1200. 1200. 1200. 1200. 1200. 1200. 1200. 1200. 1200. 1200. 1200. 1200.
1200. 1200. 1200. 1200. 1200. 1200. 1200. 1400. 1400. 1400. 1400. 1400.
1400. 1400. 1400. 1400. 1400. 1400. 1400. 1400. 1400. 1400. 1400. 1400.
1400. 1400. 1400. 1400. 1400. 1400. 1400. 1400. 1400. 1400. 1400. 1400.
1400. 1400. 1400. 1400. 1600. 1600. 1600. 1600. 1600. 1600. 1600. 1600.
1600. 1600. 1600. 1600. 1600. 1600. 1600. 1600. 1600. 1600. 1600. 1600.
1600. 1600. 1600. 1600. 1600. 1600. 1600. 1600. 1600. 1600. 1600. 1600.
1600. 1800. 1800. 1800. 1800. 1800. 1800. 1800. 1800. 1800. 1800. 1800.
1800. 1800. 1800. 1800. 1800. 1800. 1800. 1800. 1800. 1800. 1800. 1800.
1800. 1800. 1800. 1800. 1800. 1800. 1800. 1800. 1800. 1800. 2000. 2000.
2000. 2000. 2000. 2000. 2000. 2000. 2000. 2000. 2000. 2000. 2000. 2000.
2000. 2000. 2000. 2000. 2000. 2000. 2000. 2000. 2000. 2000. 2000. 2000.
2000. 2000. 2000. 2000. 2000. 2000. 2000. 2200. 2200. 2200. 2200. 2200.
2200. 2200. 2200. 2200. 2200. 2200. 2200. 2200. 2200. 2200. 2200. 2200.
2200. 2200. 2200. 2200. 2200. 2200. 2200. 2200. 2200. 2200. 2200. 2200.
2200. 2200. 2200. 2200. 2400. 2400. 2400. 2400. 2400. 2400. 2400. 2400.
2400. 2400. 2400. 2400. 2400. 2400. 2400. 2400. 2400. 2400. 2400. 2400.
2400. 2400. 2400. 2400. 2400. 2400. 2400. 2400. 2400. 2400. 2400. 2400.
2400. 2600. 2600. 2600. 2600. 2600. 2600. 2600. 2600. 2600. 2600. 2600.
2600. 2600. 2600. 2600. 2600. 2600. 2600. 2600. 2600. 2600. 2600. 2600.
2600. 2600. 2600. 2600. 2600. 2600. 2600. 2600. 2600. 2600. 2800. 2800.
2800. 2800. 2800. 2800. 2800. 2800. 2800. 2800. 2800. 2800. 2800. 2800.
2800. 2800. 2800. 2800. 2800. 2800. 2800. 2800. 2800. 2800. 2800. 2800.
2800. 2800. 2800. 2800. 2800. 2800. 2800. 3000. 3000. 3000. 3000. 3000.
3000. 3000. 3000. 3000. 3000. 3000. 3000. 3000. 3000. 3000. 3000. 3000.
3000. 3000. 3000. 3000. 3000. 3000. 3000. 3000. 3000. 3000. 3000. 3000.
3000. 3000. 3000. 3000.]
INFO:Total number of b-values 496
INFO:Number of gradient shells 15
INFO:Number of b0s 1 (b0_thr 50)
INFO:-----------------
INFO:Looking at T1.nii
INFO:-----------------
INFO:Data size (256, 256, 176)
INFO:Data type uint16
INFO:Data min 0 max 888 avg 62.940581408413976
INFO:2nd percentile 0.0 98th percentile 377.0
INFO:Native coordinate system PSR
INFO:Affine Native to RAS matrix
[[ 0. 0.01 1. -89.569]
[ -1. 0. 0. 138.451]
[ 0. 1. -0.01 -131.289]
[ 0. 0. 0. 1. ]]
INFO:Voxel size [1. 1. 1.]
INFO:---------------------------------------------------------
INFO:Looking at __DTI_AX_ep2d_2_5_iso_33d_20141015095334_4.nii
INFO:---------------------------------------------------------
INFO:Data size (96, 96, 19, 496)
INFO:Data type int16
INFO:Data min 0 max 1257 avg 58.62918037280702 of vol 0
INFO:2nd percentile 0.0 98th percentile 234.0 of vol 0
INFO:Native coordinate system LAS
INFO:Affine Native to RAS matrix
[[ -2.498 0.084 0.067 113.641]
[ 0.069 2.451 -0.488 -104.142]
[ 0.082 0.486 2.451 -31.504]
[ 0. 0. 0. 1. ]]
INFO:Voxel size [2.5 2.5 2.5]
We can visualize the data using dipy_horizon
dipy_horizon dwi.nii

Visualization of a slice from the first volume of the diffusion data#
We can use dipy_median_otsu to build a brain mask for the diffusion data:
dipy_median_otsu dwi.nii --median_radius 2 --numpass 1 --vol_idx 10-50 --out_dir out_work
Visualize the mask using dipy_horizon:
dipy_horizon out_work/brain_mask.nii.gz

Visualization of a slice from the generated brain mask#
Perform DTI using dipy_fit_dti. The input of this function is the DWI data, b-values and b-vector files and the
brain mask that we calculated in the previous step:
dipy_fit_dti dwi.nii dwi.bval dwi.bvec out_work/brain_mask.nii.gz --out_dir out_work/
The default options of the script generate the following files ad.nii.gz, evecs.nii.gz, md.nii.gz, rgb.nii.gz, fa.nii.gz, mode.nii.gz, tensors.nii.gz, evals.nii.gz, ga.nii.gz and rd.nii.gz.
Visualize DEC map:
dipy_horizon out_work/rgb.nii.gz --rgb
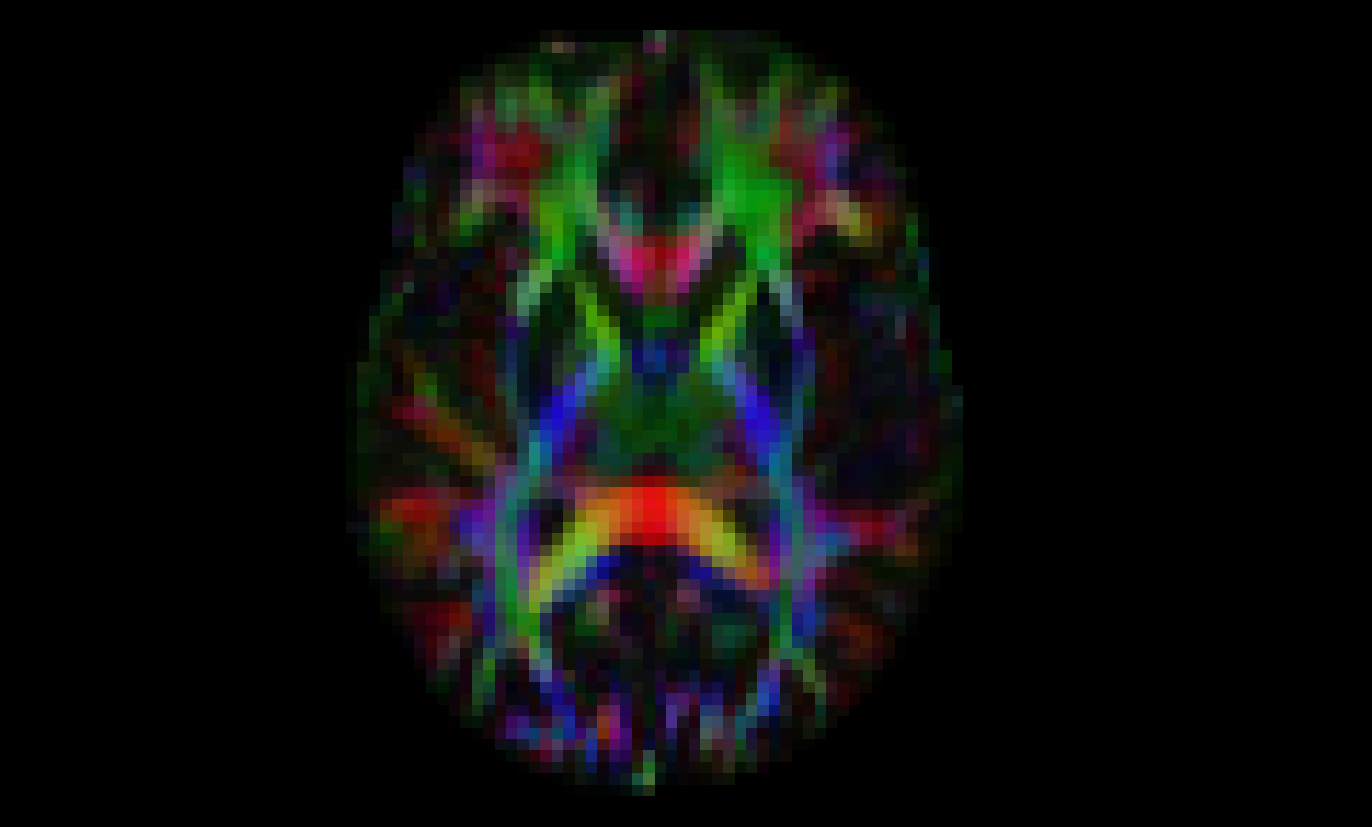
Visualization of a slice from the first volume of DEC image#
We can now move to more advanced reconstruction models. One of the fastest we can use is Constant Solid Angle (CSA) [3]
dipy_fit_csa dwi.nii dwi.bval dwi.bvec out_work/brain_mask.nii.gz --out_dir out_work/
Now, to move into doing some tracking we will need some seeds. We can generate seeds in the following way
dipy_mask out_work/fa.nii.gz 0.4 --out_dir out_work/ --out_mask seed_mask.nii.gz
Build tractography with the peaks.pam5 file as input using the fast EuDX algorithm
[4]
dipy_track out_work/peaks.pam5 out_work/fa.nii.gz out_work/seed_mask.nii.gz --out_dir out_work/ --out_tractogram tracks_from_peaks.trk --tracking_method eudx
We can visualize the result using dipy_horizon:
dipy_horizon out_work/tracks_from_peaks.trk

Showing tracks from the specific dataset. This dataset contains only a few slices.#
For more information about each command line, try calling the -h flag for example
dipy_horizon -h
should provide the available options
usage: dipy_horizon [-h] [--cluster] [--cluster_thr float] [--random_colors]
[--length_gt float] [--length_lt float]
[--clusters_gt int] [--clusters_lt int] [--native_coords]
[--stealth] [--emergency_header str]
[--bg_color [float [float ...]]]
[--disable_order_transparency] [--out_dir str]
[--out_stealth_png str] [--force] [--version]
[--out_strat string] [--mix_names] [--log_level string]
[--log_file string]
input_files [input_files ...]
Otherwise please see Command Line Utilities Reference.
The commands shown in this tutorial are not by any stretch of imagination what we propose as a complete solution to tracking but a mere introduction to DIPY’s command interfaces. Medical imaging requires a number of steps that depend on the goal of the analysis strategy.
Nonetheless, if you are using these commands do cite the relevant papers to support the DIPY developers so that they can continue maintaining and extending these tools.