Gibbs Unringing#
This tutorial walks through the steps to perform Gibbs unringing using DIPY.
You can try this using your own data; we will be using the data in DIPY. You can check how to fetch the DIPY data.
Suppress Gibbs Oscillations#
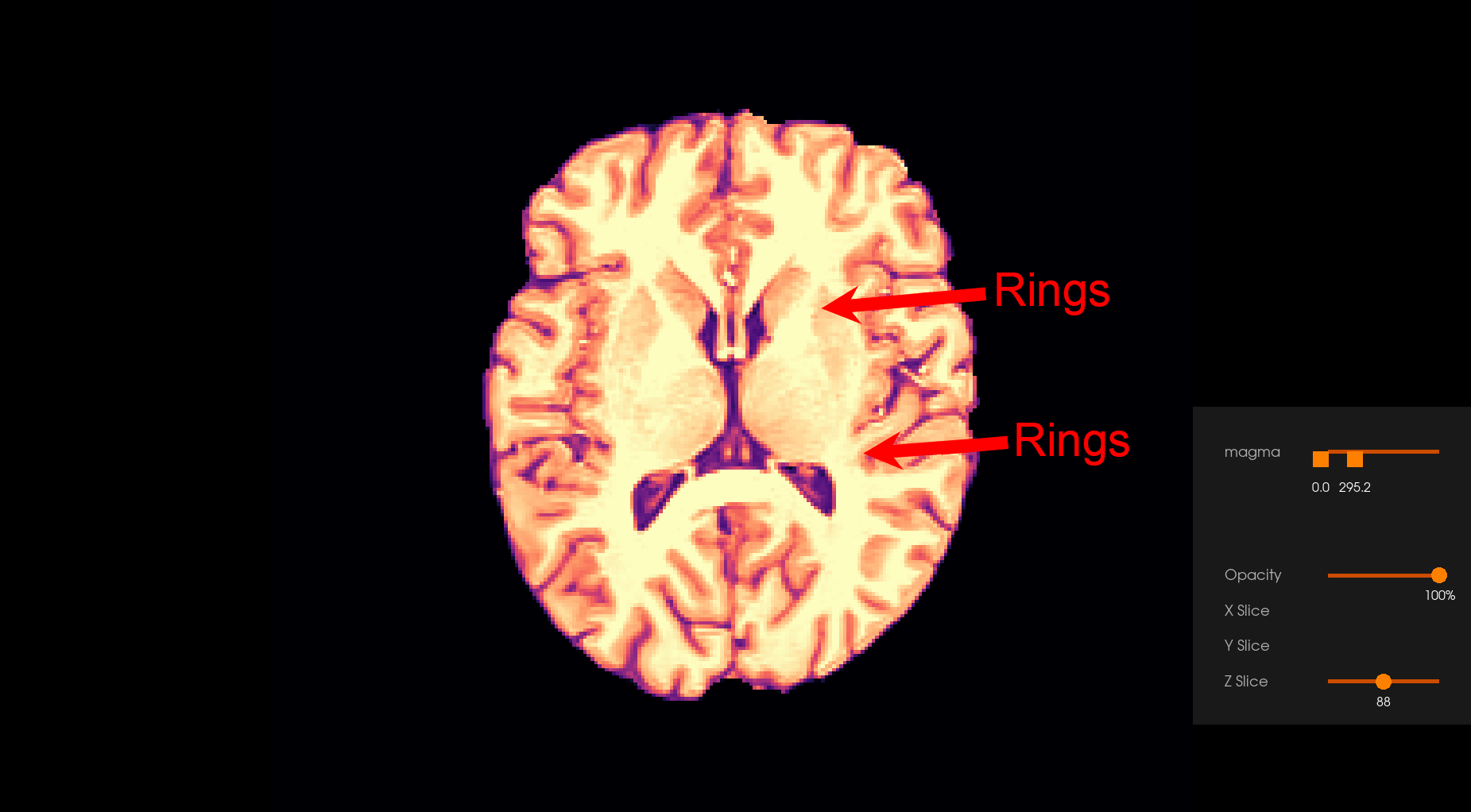
Magnetic Resonance (MR) images are reconstructed from the Fourier coefficients of acquired k-space images. Since only a finite number of Fourier coefficients can be acquired in practice, reconstructed MR images can be corrupted by Gibbs artefacts, which is manifested by intensity oscillations adjacent to edges of different tissue types [1]. Although this artefact affects MR images in general, in the context of diffusion-weighted imaging, Gibbs oscillations can be magnified in derived diffusion-based estimates [1], [2].
We will use the tissue_data dataset in DIPY to showcase the ability to
remove Gibbs ringing artefacts. As with any other workflow in DIPY, you can
also use your own data!
You may want to create an output directory to save the resulting Gibbs ringing-free volume:
mkdir gibbs_ringing_output
To run the Gibbs unringing method, we need to specify the path to the input
data. This path may contain wildcards to process multiple inputs at once.
You can also specify the optional arguments. In this case, we will be
specifying the number of processes (num_processes) and output directory
(out_dir). The number of processes allows one to exploit the available
computational power and accelerate the processing. The maximum number of
processes available depends on the CPU of the computer, so users are expected
to set an appropriate value based on their platform. Set num_processes to
-1 to use all available cores.
To run the Gibbs unringing on the data it suffices to execute the
dipy_gibbs_ringing command, e.g.:
dipy_gibbs_ringing data/tissue_data/t1_brain_denoised.nii.gz --num_processes 4 --out_dir "gibbs_ringing_output"
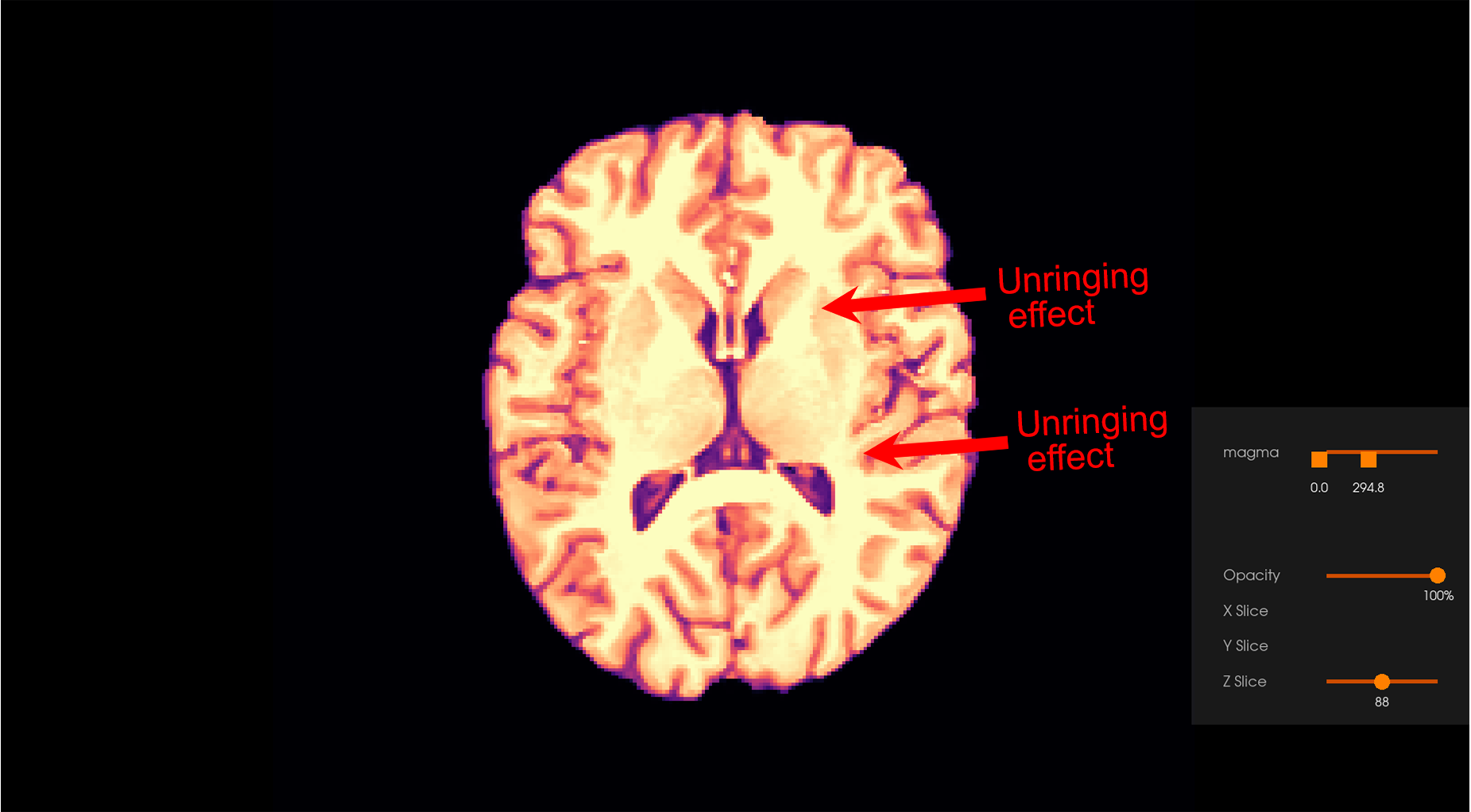
This command will apply the Gibbs unringing procedure to the input MR image
and write the artefact-free result to the gibbs_ringing_output directory.
In case no output directory is specified, the Gibbs artefact-free output volume is saved to the current directory by default.
Note: Users are recommended to zoom on each image by clicking on them to see the Gibbs unringing effect.
Before Gibbs unringing |
After Gibbs unringing |
|---|---|
|
|