Denoising#
This tutorial walks through the steps to denoise diffusion-weighted MR images using DIPY. Multiple denoising methods are available in DIPY.
You can try these methods using your own data; we will be using the data in DIPY. You can check how to fetch the DIPY data.
Denoising using Overcomplete Local PCA#
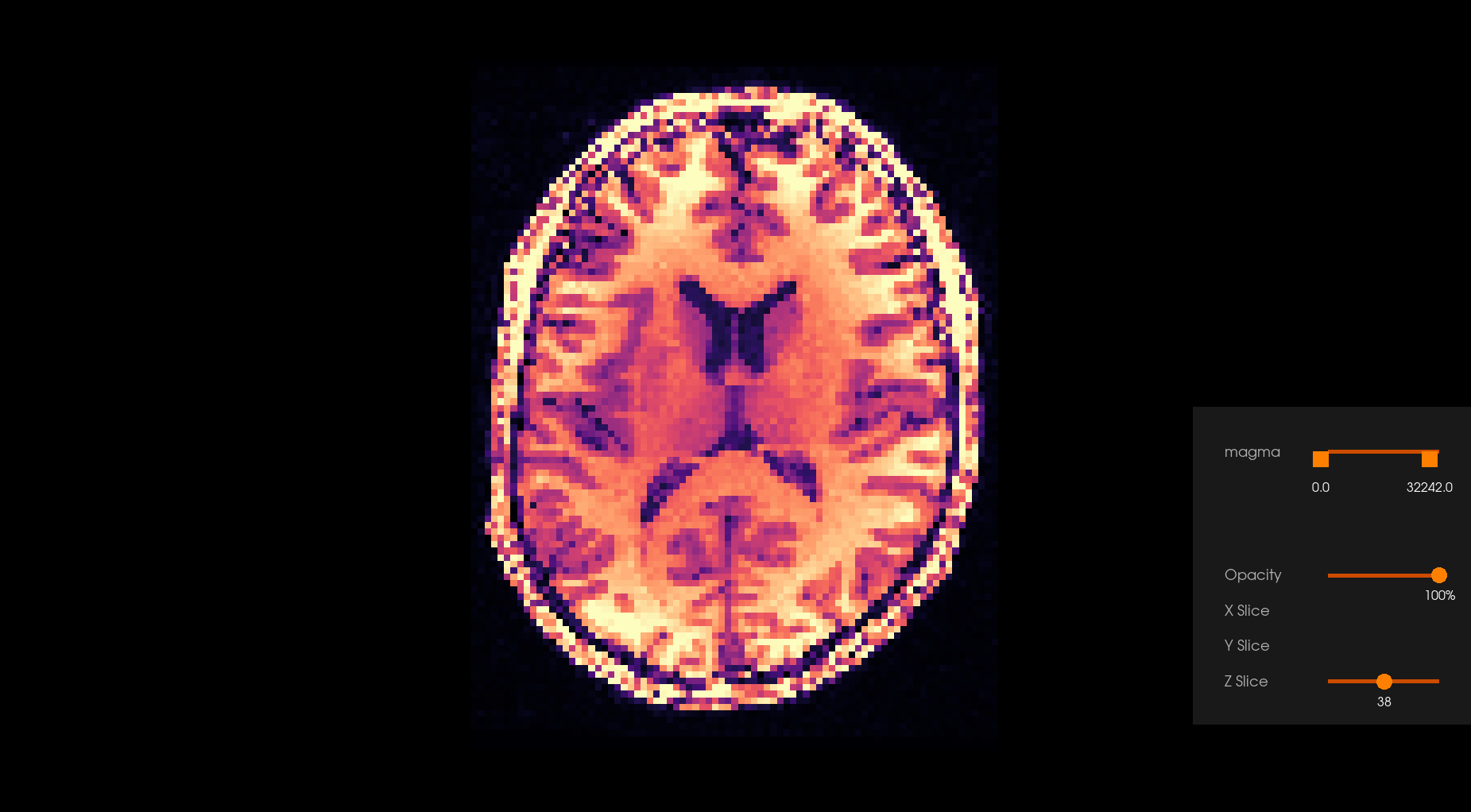
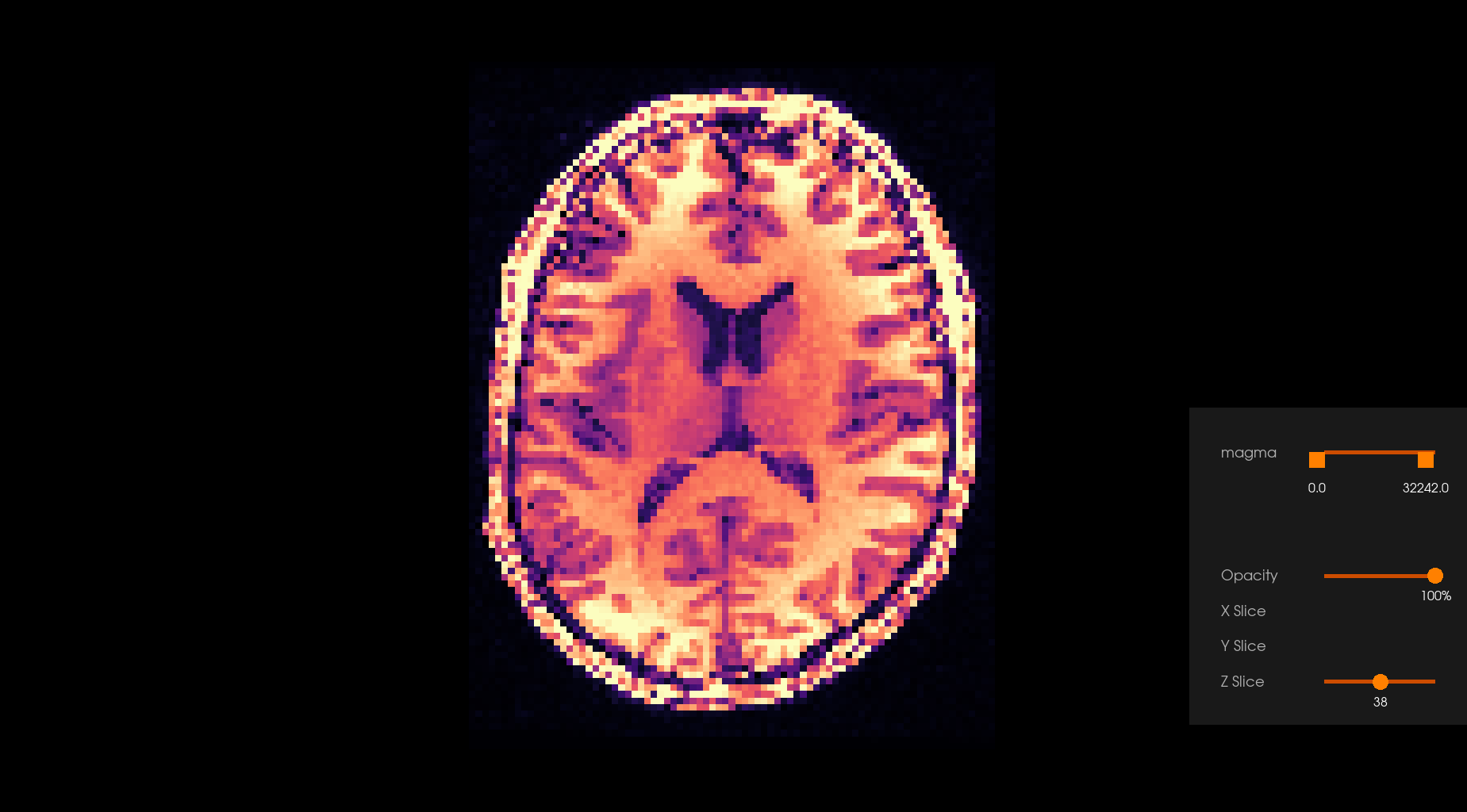
Denoising algorithms based on principal components analysis (PCA) are effective denoising methods because they explore the redundancy of the multi-dimensional information of diffusion-weighted datasets. The basic idea behind the PCA-based denoising algorithms is to perform a low-rank approximation by thresholding the eigenspectrum of the noisy signal matrix.
The algorithm to perform an Overcomplete Local PCA-based (LPCA) denoising involves the following steps:
Estimating the local noise variance at each voxel.
Applying a PCA in local patches around each voxel over the gradient directions.
Thresholding the eigenvalues based on the local estimate of the noise variance, and then doing a PCA reconstruction.
The Overcomplete Local PCA algorithm turns out to work well on diffusion MRI owing to the 4D structure of DWI acquisitions where the q-space is typically oversampled giving highly correlated 3D volumes of the same subject.
For illustrative purposes of the Overcomplete Local PCA denoising method, we
will be using the stanford_hardi dataset, but you can also use your own
data.
The workflow for the LPCA denoising requires the paths to the diffusion input file, as well as the b-values and b-vectors files.
You may want to create an output directory to save the denoised data, e.g.:
mkdir denoise_lpca_output
To run the Overcomplete Local PCA denoising on the data it suffices to execute
the dipy_denoise_lpca command, e.g.:
dipy_denoise_lpca data/stanford_hardi/HARDI150.nii.gz data/stanford_hardi/HARDI150.bval data/stanford_hardi/HARDI150.bvec --out_dir "denoise_lpca_output"
This command will denoise the diffusion image and save it to the
denoise_lpca_output directory, defaulting the resulting image file name to
dwi_lpca.nii.gz. In case the output directory (out_dir) parameter is not
specified, the denoised diffusion image will be saved to the current directory
by default.
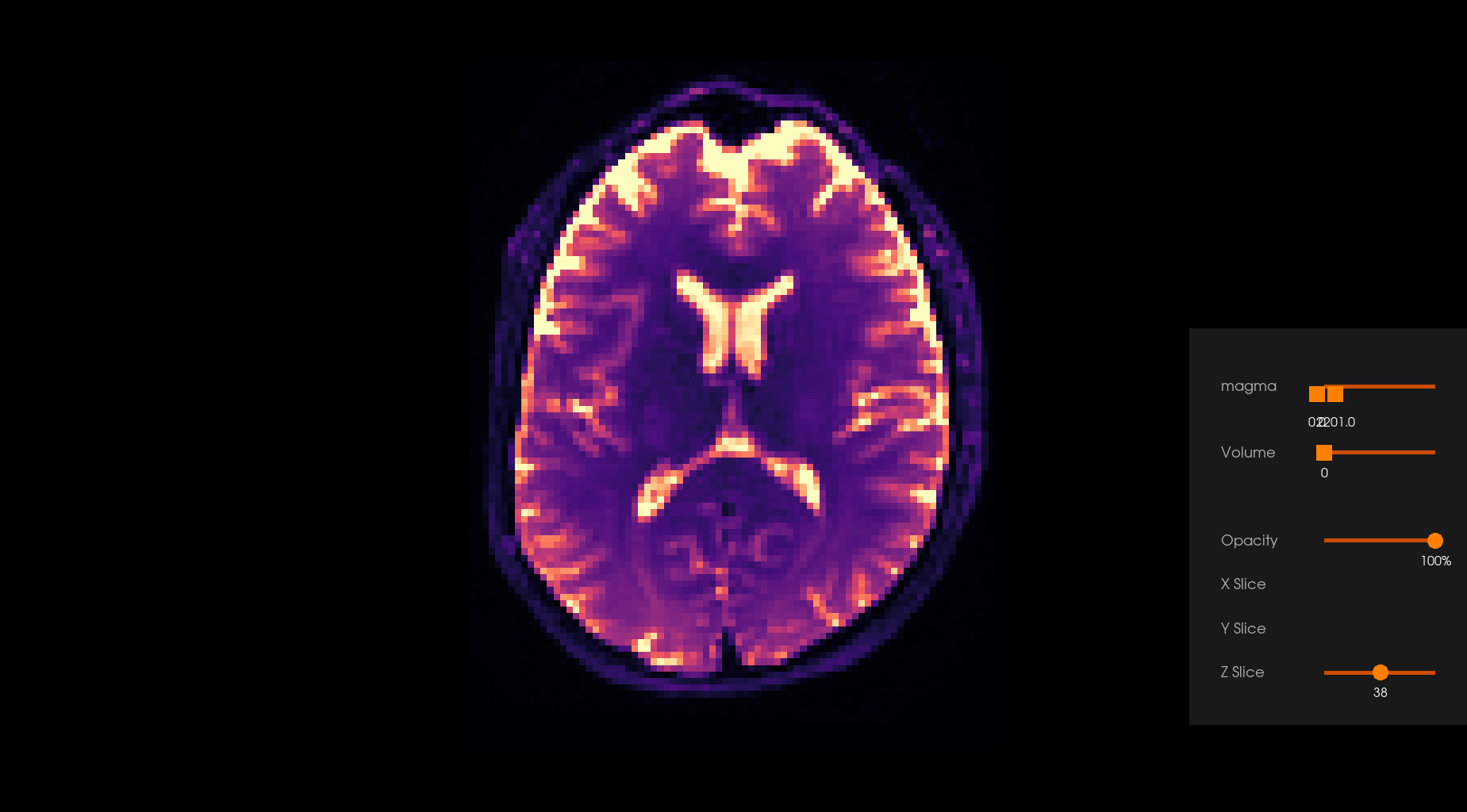
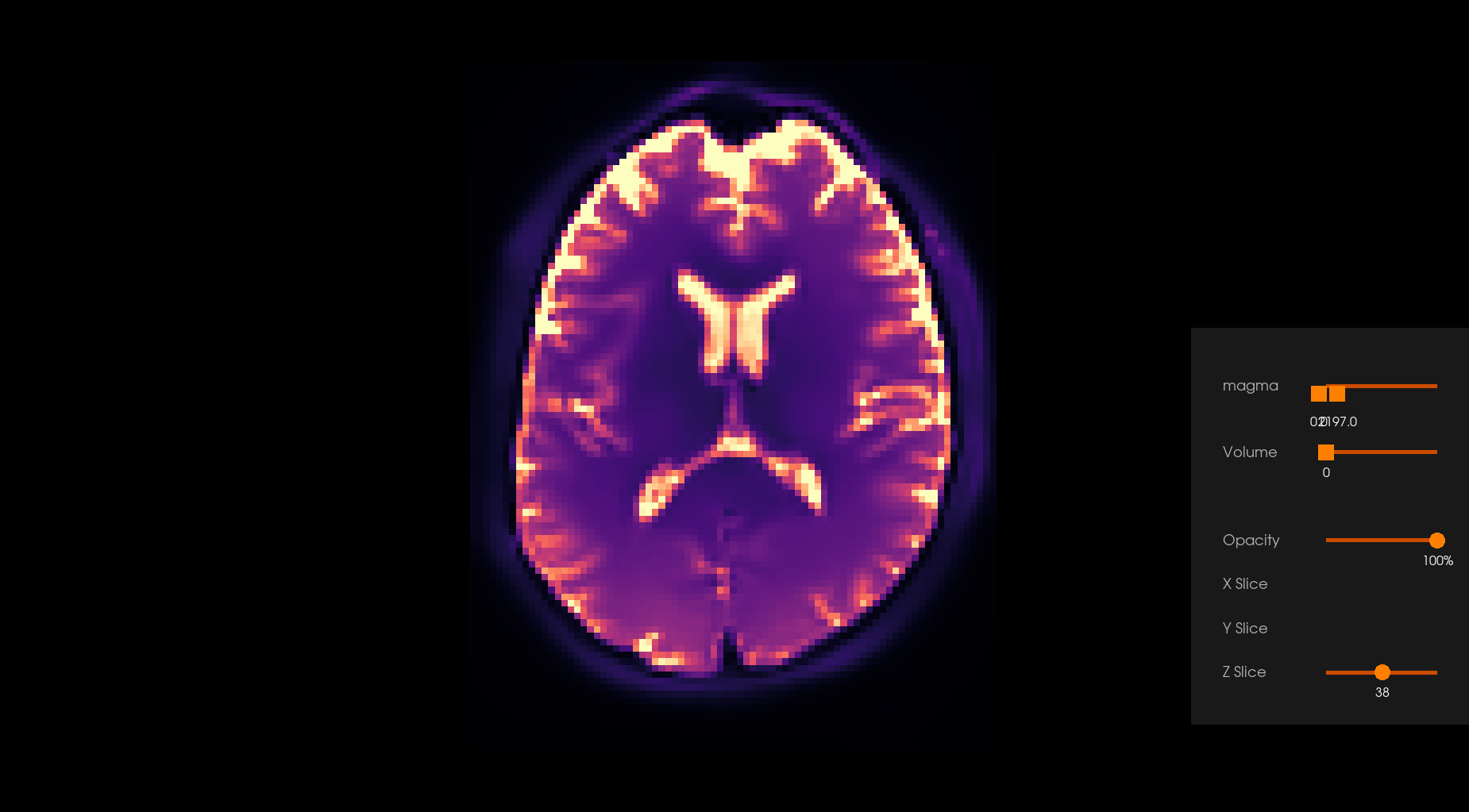
Note: Depending on the parameters’ values, the effect of the denoising can be subtle or even hardly noticeable, apparent or visible, depending on the choice. Users are encouraged to carefully choose the parameters.
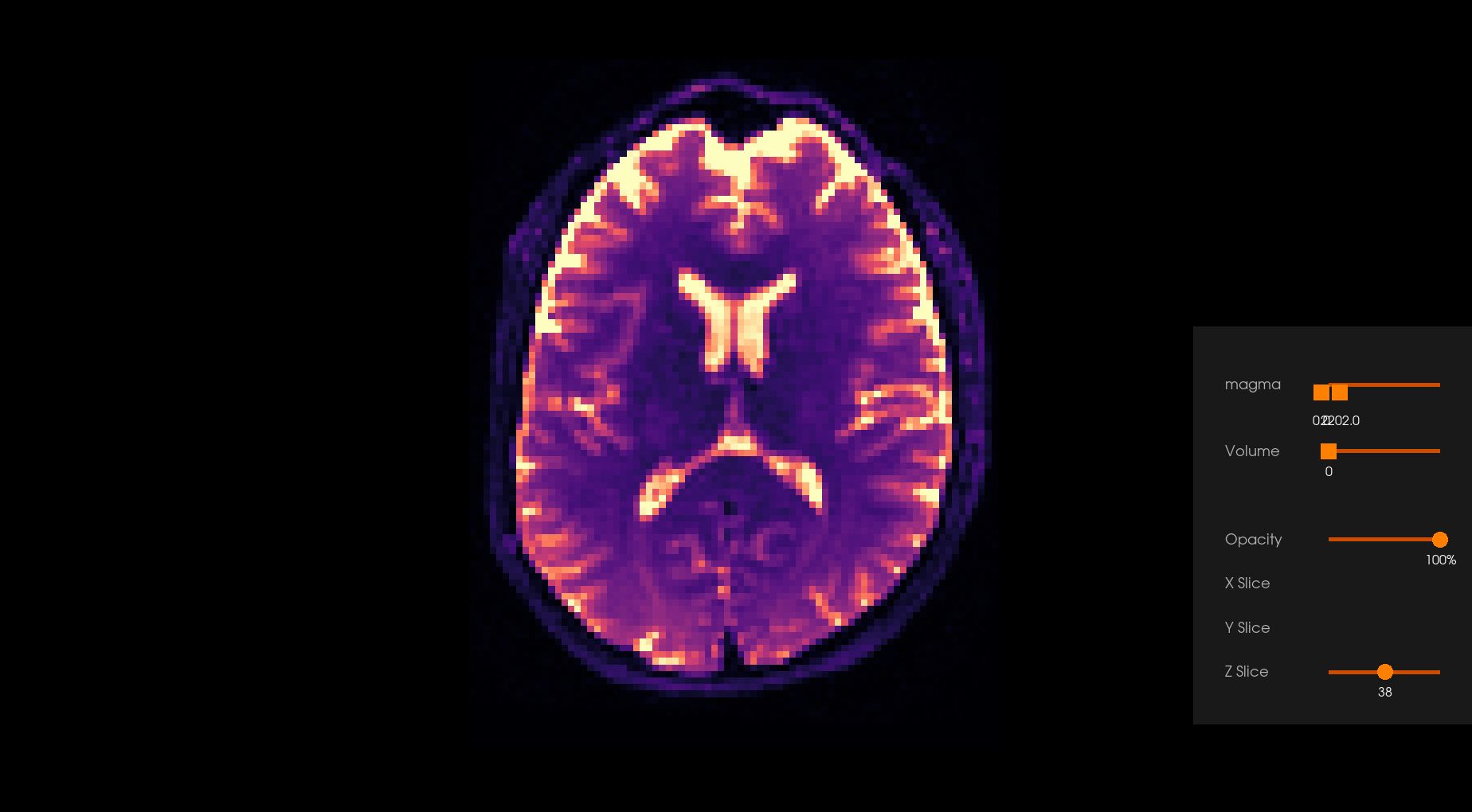
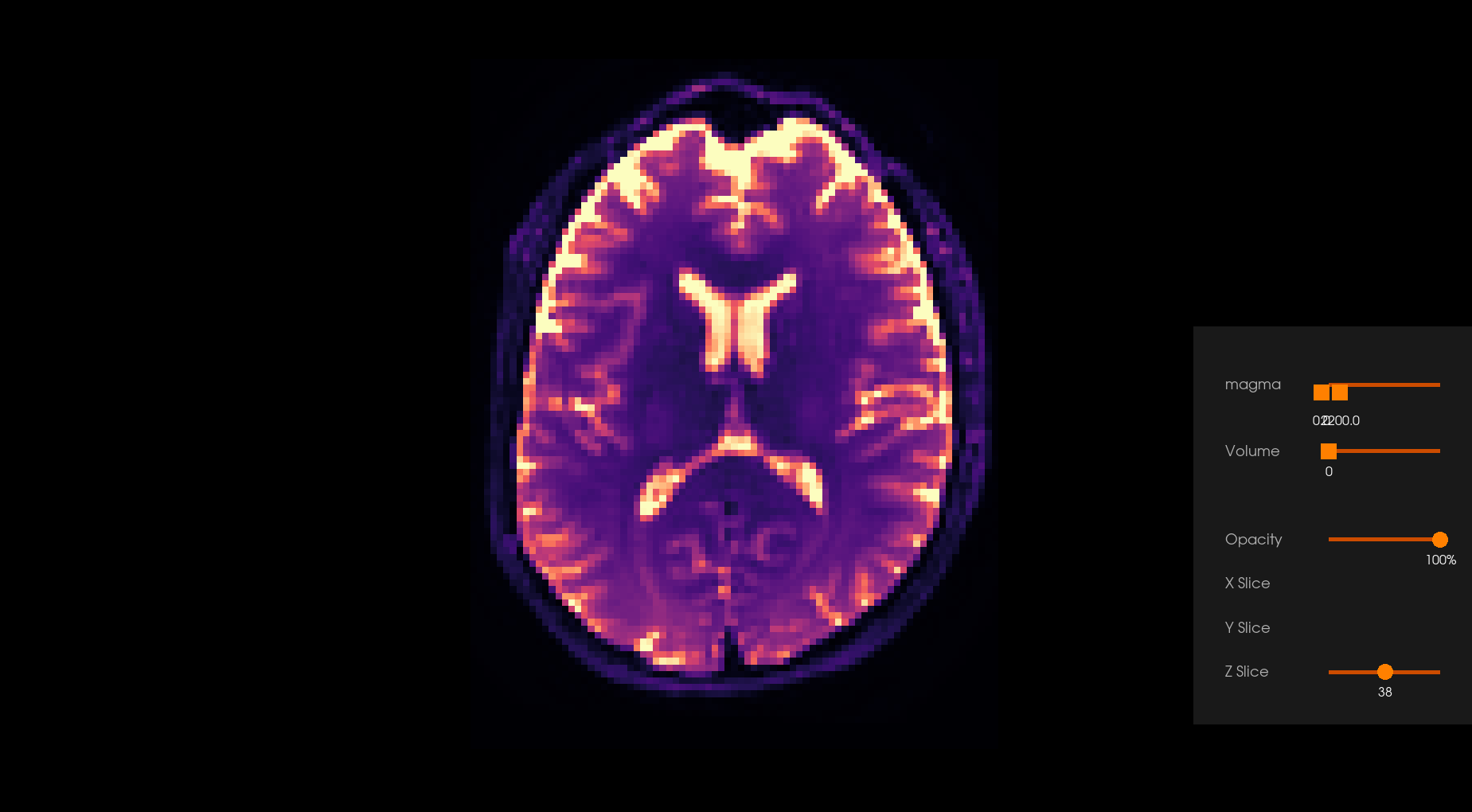
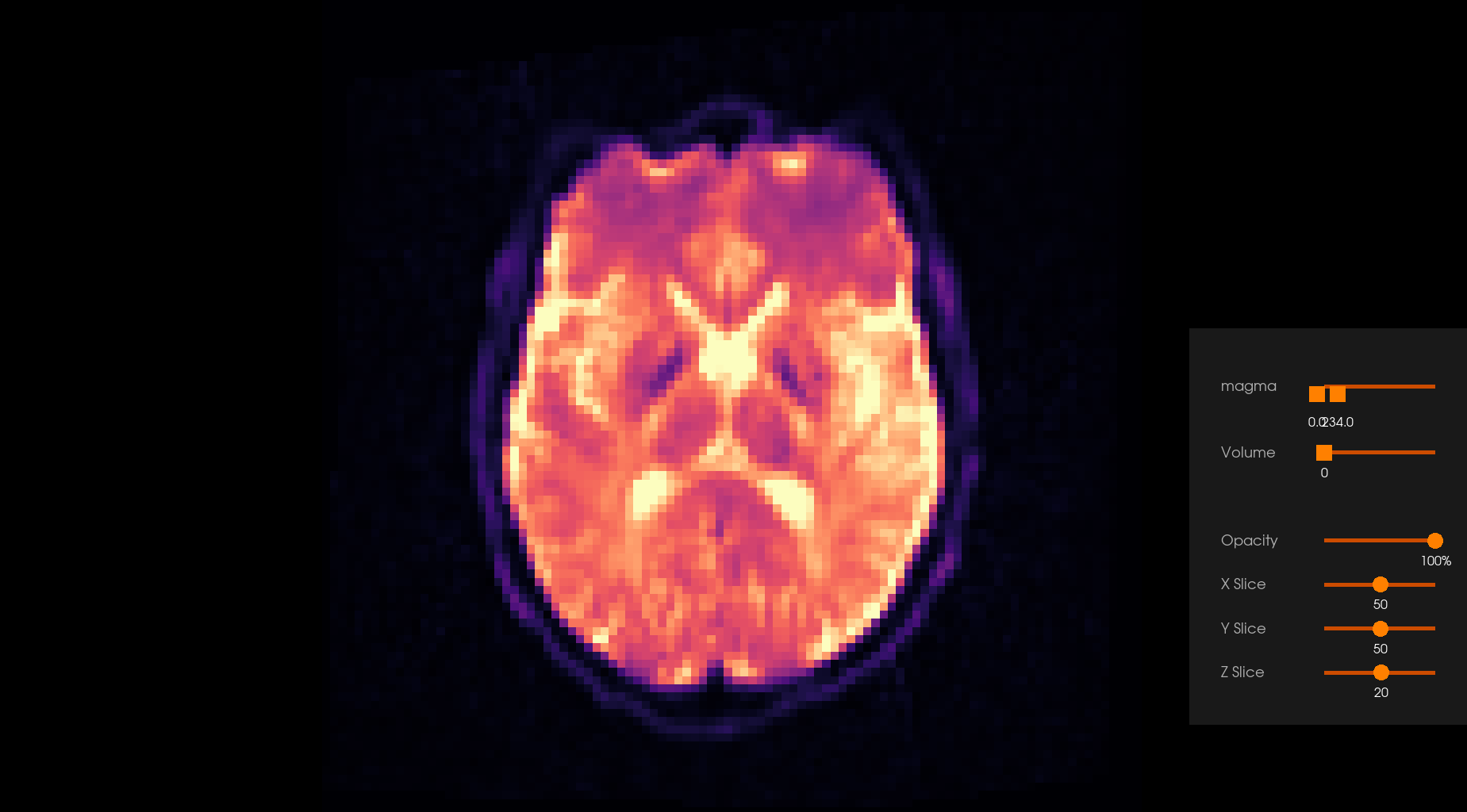
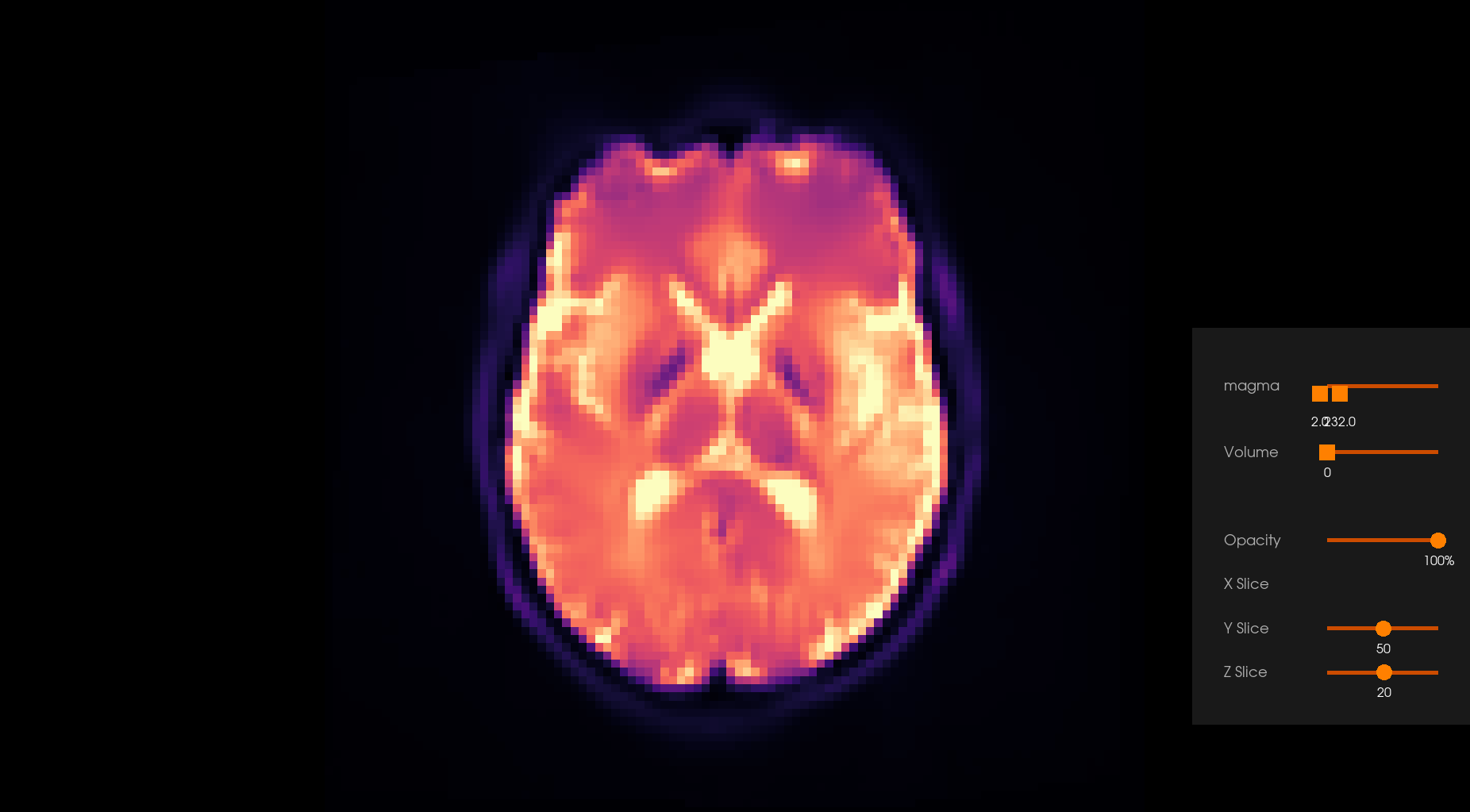
Before Denoising |
After Denoising |
|---|---|
|
|
Denoising using Marcenko-Pastur PCA#
The principal components classification can be performed based on prior noise variance estimates or automatically based on the Marcenko-Pastur distribution. In addition to noise suppression, the Marcenko-Pastur PCA (MPPCA) algorithm can be used to get the standard deviation of the noise.
We will use the sherbrooke_3shell dataset in DIPY to showcase this denoising
method. As with any other workflow in DIPY, you can also use your own data!
We will create a directory where to save the denoised image (e.g.:
denoise_mppca_output):
mkdir denoise_mppca_output
In order to run the MPPCA denoising method, we need to specify the location of the diffusion data file, followed by the optional arguments. In this case, we will be specifying the patch radius value and the output directory.
The MPPCA denoising method is run using the dipy_denoise_mppca command,
e.g.:
dipy_denoise_mppca data/sherbrooke_3shell/HARDI193.nii.gz --patch_radius 10 --out_dir "denoise_mppca_output"
This command will denoise the diffusion image and save it to the specified output directory.
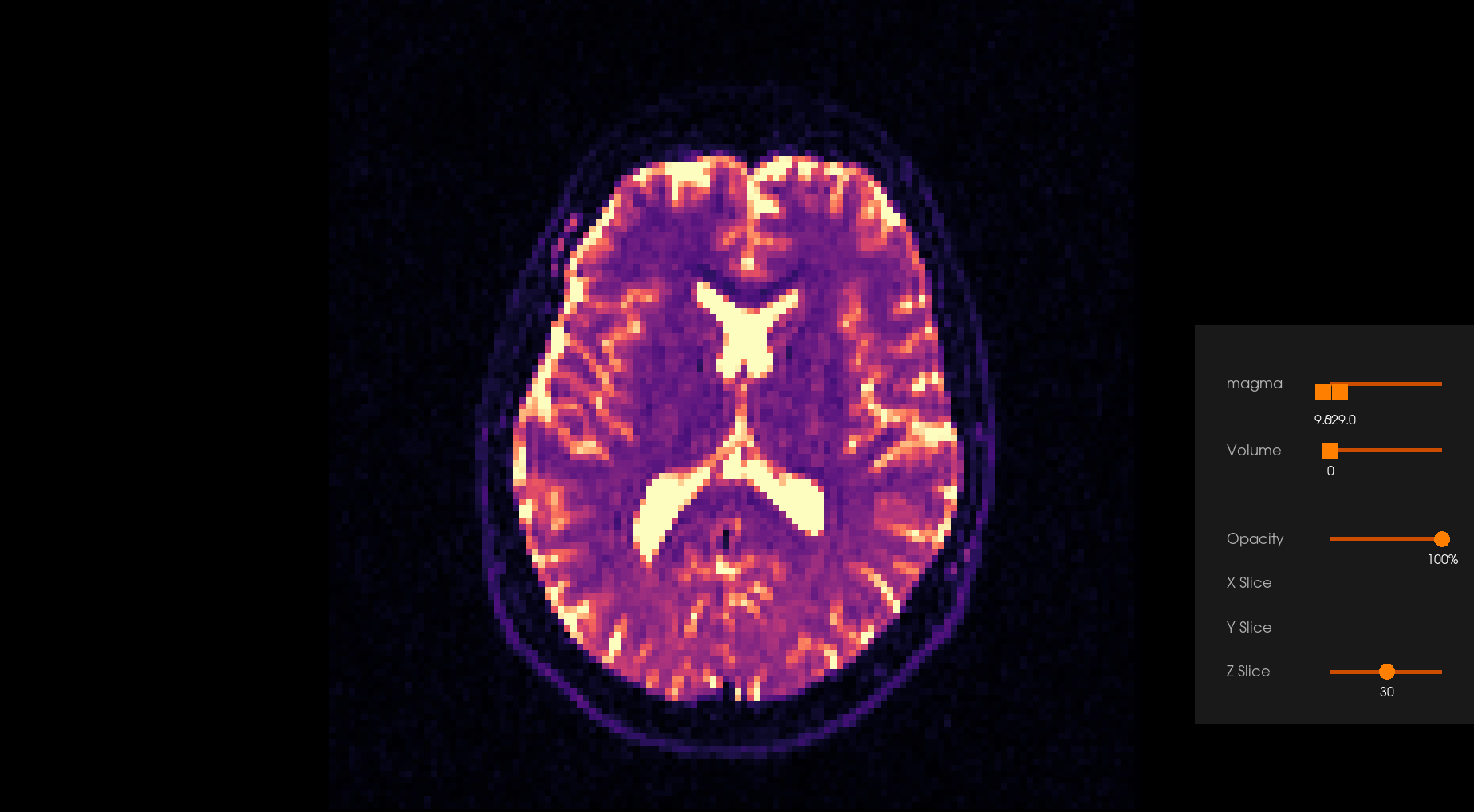
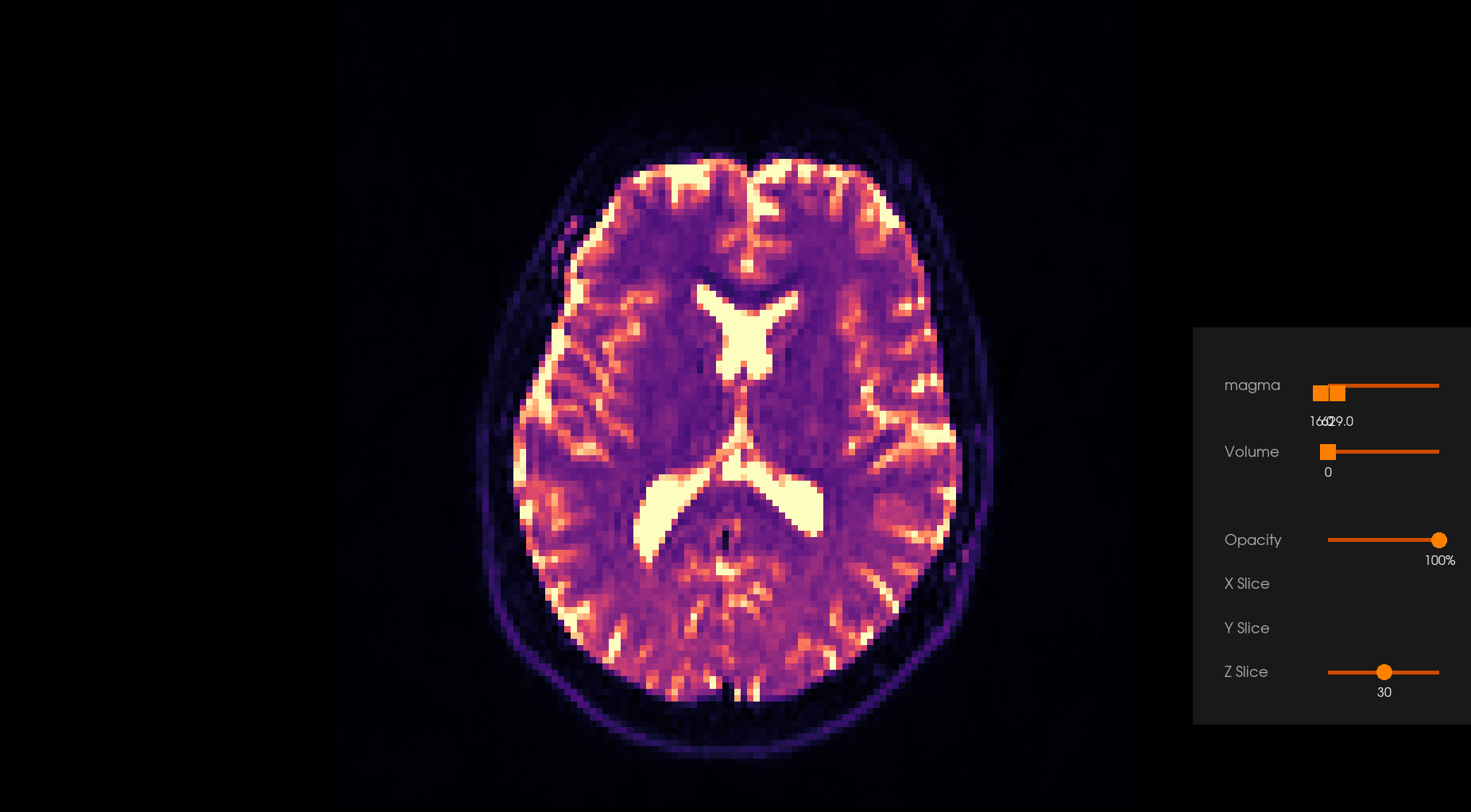
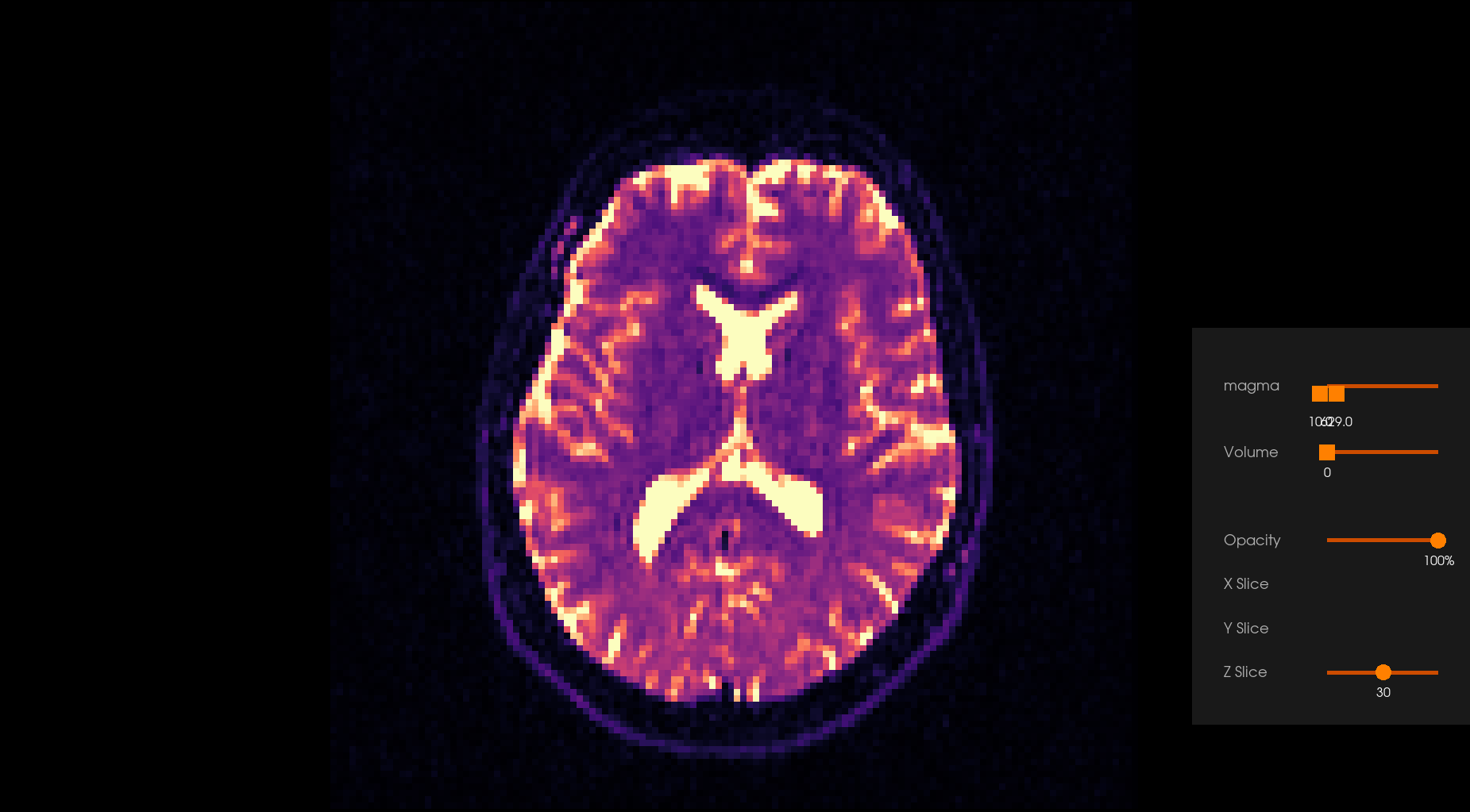
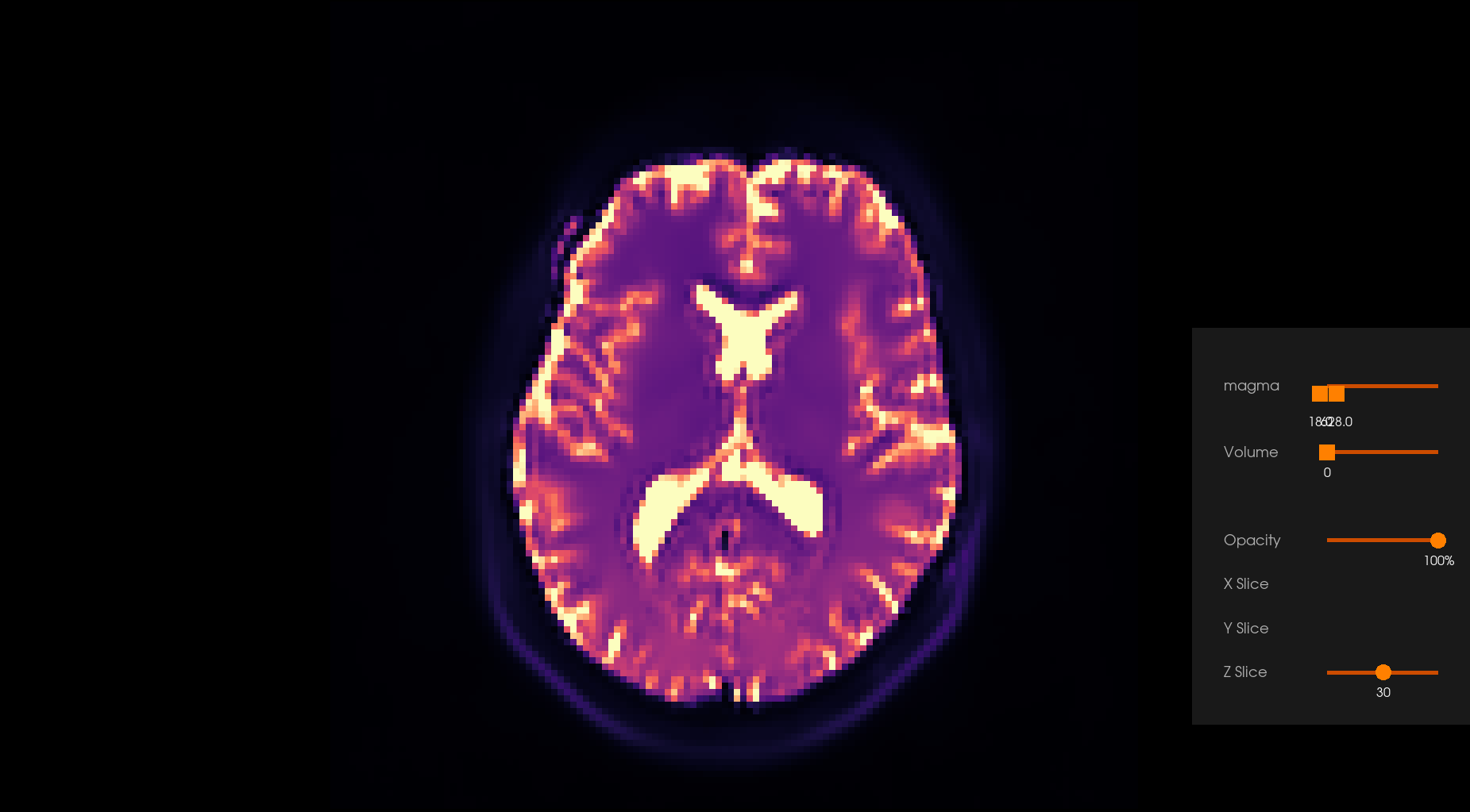
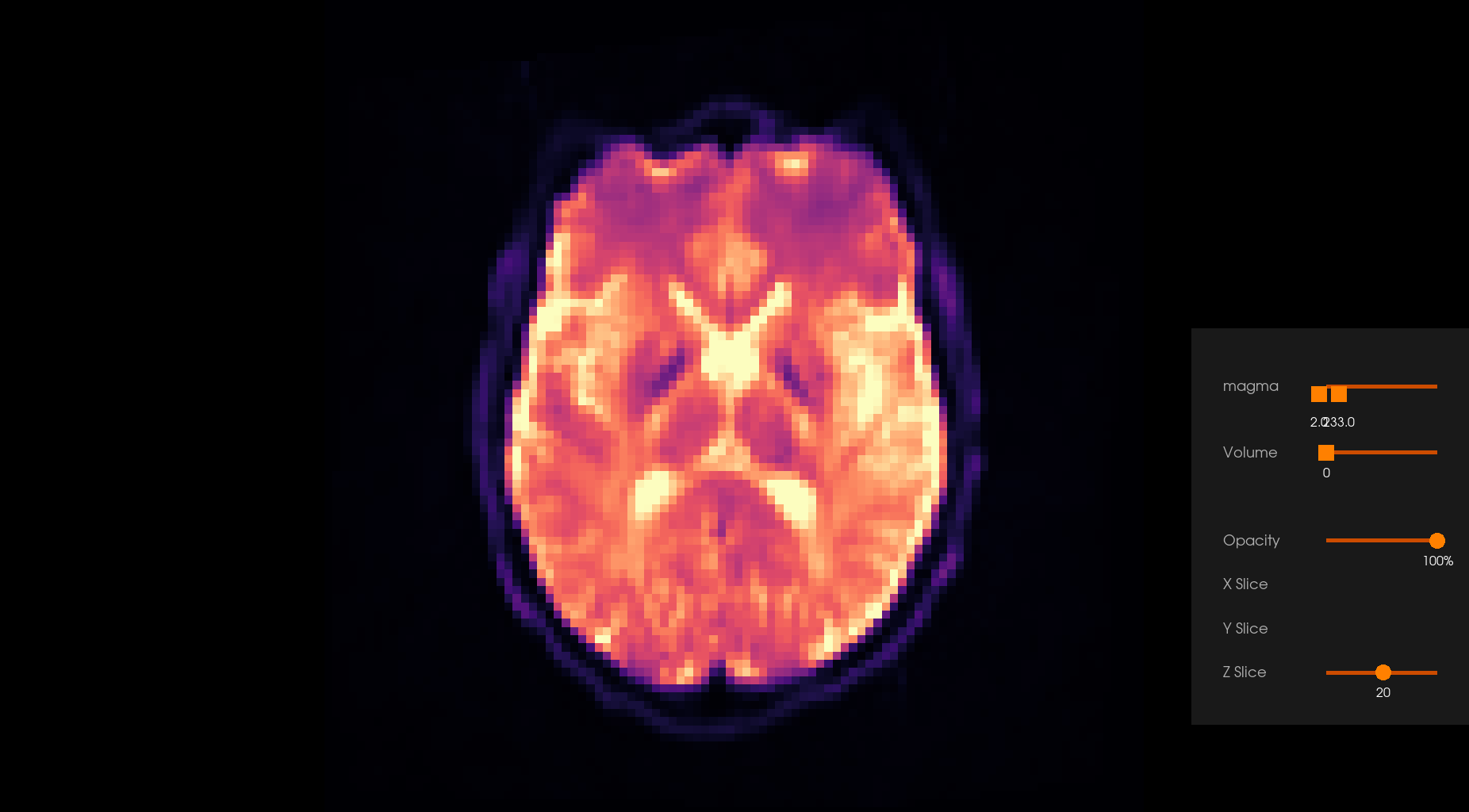
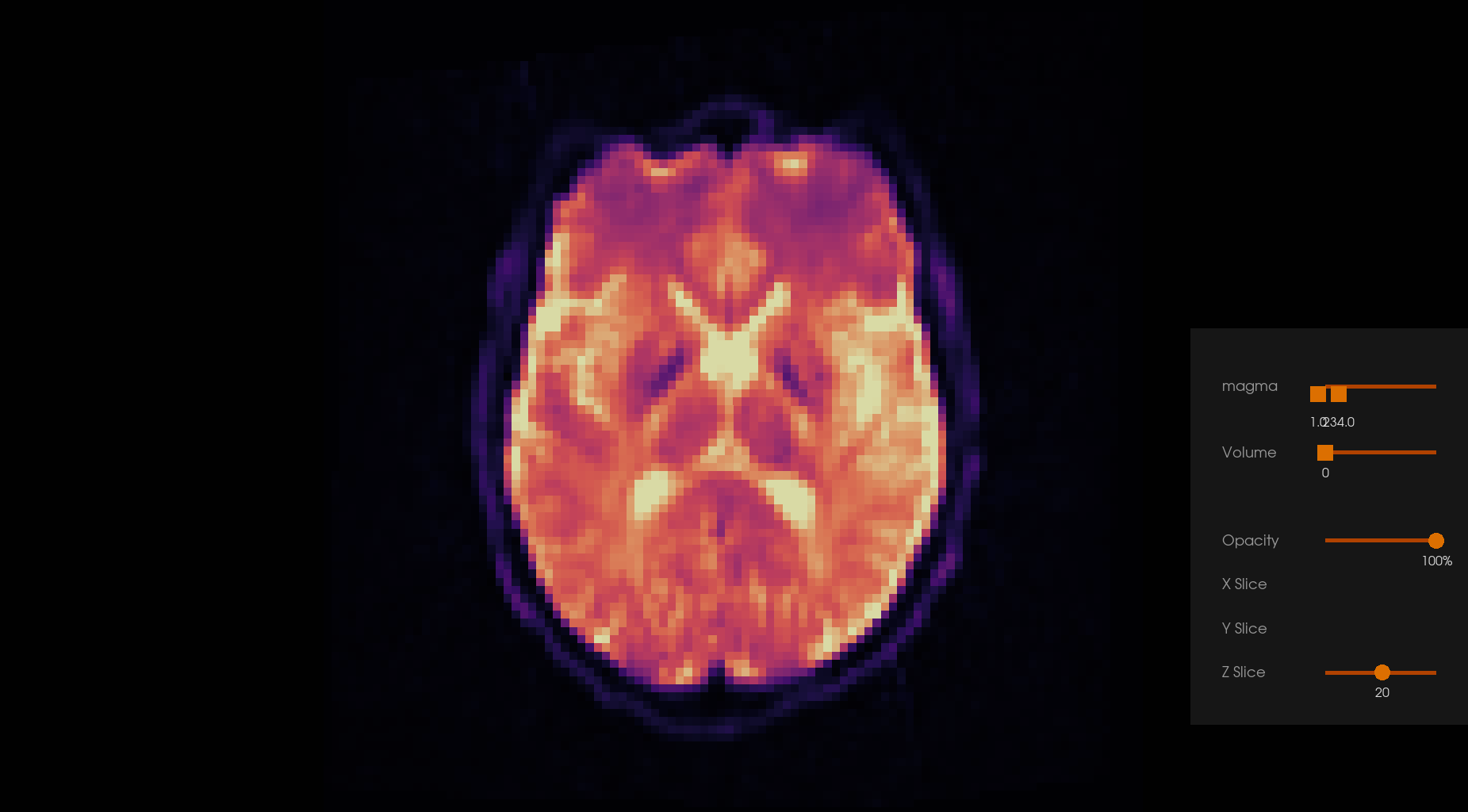
Before Denoising |
After Denoising |
|---|---|
|
|
Denoising using NLMEANS#
In the Non-Local Means algorithm (NLMEANS) [1], [2], the value of a pixel is replaced by an average of a set of other pixel values: the specific patches centered on the other pixels are contrasted to the patch centered on the pixel of interest, and the average only applies to pixels with patches close to the current patch. This algorithm can also restore good textures, which are distorted by other denoising algorithms.
The Non-Local Means method can be used to denoise \(N\)-D image data (i.e. 2D, 3D, 4D, etc.), and thus enhance their SNR.
We will use the cfin_multib dataset in DIPY to showcase this denoising
method. As with any other workflow in DIPY, you can also use your own data!
In order to run the NLMEANS denoising method, we need to specify the location of the
diffusion data file, followed by the optional arguments. In this case, we will be
specifying the noise standard deviation estimate (sigma) and patch radius
values, and the output directory.
We will create a directory where to save the denoised image (e.g.:
denoise_nlmeans_output):
mkdir denoise_nlmeans_output
The NLMEANS denoising is performed using the dipy_denoise_nlmeans command,
e.g.:
dipy_denoise_nlmeans data/cfin_multib/__DTI_AX_ep2d_2_5_iso_33d_20141015095334_4.nii --sigma 2 --patch_radius 2 --out_dir "denoise_nlmeans_output"
The command will denoise the input diffusion volume and write the result to the specified output directory.
Before Denoising |
After Denoising |
|---|---|
|
|
Overview of Denoising Methods#
Note: Users are recommended to zoom (click on each image) to see the denoising effect.
Diffusion#
Dataset |
Original Image |
Denoise LCPA |
Denoise MPPCA |
Denoise NLMEANS |
|---|---|---|---|---|
sherbrooke_3shell |
|
|
|
|
stanford_hardi |
|
|
|
|
cfin_multib |
|
|
|
|
Structural#
Dataset |
Original Image |
Denoise NLMEANS |
|---|---|---|
stanford_hardi T1 |
|
|