Mean signal diffusion kurtosis imaging (MSDKI)¶
Several microstructural models have been proposed to increase the specificity of diffusion-weighted data; however, improper model assumptions are known to compromise the validity of the model’s estimates [NetoHe2019]. To avoid misleading interpretation, it might be enough to characterize diffusion-weighted data using signal representation techniques. For example, assuming that the degree of non-Gaussian diffusion decreases with tissue degeneration, this can be sensitive to general microstructural alterations. Although this cannot be used to distinguish different mechanisms of microstructural changes (e.g. axonal loss vs demyelination), the degree of non-Gaussian diffusion can provide insights on the general condition of tissue microstructure and provide useful markers to understanding, for instance, the relationship between brain microstructure changes and alterations in behaviour (e.g. [Price2017]).
Diffusion Kurtosis Imaging is one of the conventional ways to estimate the degree of non-Gaussian diffusion (see Reconstruction of the diffusion signal with the kurtosis tensor model). However, as previously pointed [NetoHe2015], standard kurtosis measures do not only depend on microstructural properties but also on mesoscopic properties such as fiber dispersion or the intersection angle of crossing fibers.
In the following example, we show how one can process the diffusion kurtosis from mean signals (also known as powder-averaged signals) and obtain a characterization of non-Gaussian diffusion independently to the degree of fiber organization [NetoHe2018]. In the first part of this example, the properties of the measures obtained from the mean signal diffusion kurtosis imaging [NetoHe2018] are illustrated using synthetic data. Secondly, the mean signal diffusion kurtosis imaging will be applied to in-vivo MRI data.
Let’s import all relevant modules:
import numpy as np
import matplotlib.pyplot as plt
# Reconstruction modules
import dipy.reconst.dki as dki
import dipy.reconst.msdki as msdki
# For simulations
from dipy.sims.voxel import multi_tensor
from dipy.core.gradients import gradient_table
from dipy.core.sphere import disperse_charges, HemiSphere
# For in-vivo data
from dipy.data import fetch_cfin_multib
from dipy.data import read_cfin_dwi
from dipy.segment.mask import median_otsu
Testing MSDKI in synthetic data¶
We simulate representative diffusion-weighted signals using MultiTensor simulations (for more information on this type of simulations see MultiTensor Simulation). For this example, simulations are produced based on the sum of four diffusion tensors to represent the intra- and extra-cellular spaces of two fiber populations. The parameters of these tensors are adjusted according to [NetoHe2015] (see also DKI MultiTensor Simulation).
mevals = np.array([[0.00099, 0, 0],
[0.00226, 0.00087, 0.00087],
[0.00099, 0, 0],
[0.00226, 0.00087, 0.00087]])
For the acquisition parameters of the synthetic data, we use 60 gradient directions for two non-zero b-values (1000 and 2000 \(s/mm^{2}\)) and two zero bvalues (note that, such as the standard DKI, MSDKI requires at least three different b-values).
# Sample the spherical coordinates of 60 random diffusion-weighted directions.
n_pts = 60
theta = np.pi * np.random.rand(n_pts)
phi = 2 * np.pi * np.random.rand(n_pts)
# Convert direction to cartesian coordinates.
hsph_initial = HemiSphere(theta=theta, phi=phi)
# Evenly distribute the 60 directions
hsph_updated, potential = disperse_charges(hsph_initial, 5000)
directions = hsph_updated.vertices
# Reconstruct acquisition parameters for 2 non-zero=b-values and 2 b0s
bvals = np.hstack((np.zeros(2), 1000 * np.ones(n_pts), 2000 * np.ones(n_pts)))
bvecs = np.vstack((np.zeros((2, 3)), directions, directions))
gtab = gradient_table(bvals, bvecs)
Simulations are looped for different intra- and extra-cellular water volume fractions and different intersection angles of the two-fiber populations.
# Array containing the intra-cellular volume fractions tested
f = np.linspace(20, 80.0, num=7)
# Array containing the intersection angle
ang = np.linspace(0, 90.0, num=91)
# Matrix where synthetic signals will be stored
dwi = np.empty((f.size, ang.size, bvals.size))
for f_i in range(f.size):
# estimating volume fractions for individual tensors
fractions = np.array([100 - f[f_i], f[f_i], 100 - f[f_i], f[f_i]]) * 0.5
for a_i in range(ang.size):
# defining the directions for individual tensors
angles = [(ang[a_i], 0.0), (ang[a_i], 0.0), (0.0, 0.0), (0.0, 0.0)]
# producing signals using Dipy's function multi_tensor
signal, sticks = multi_tensor(gtab, mevals, S0=100, angles=angles,
fractions=fractions, snr=None)
dwi[f_i, a_i, :] = signal
Now that all synthetic signals were produced, we can go forward with MSDKI fitting. As other Dipy’s reconstruction techniques, the MSDKI model has to be first defined for the specific GradientTable object of the synthetic data. For MSDKI, this is done by instantiating the MeanDiffusionKurtosisModel object in the following way:
msdki_model = msdki.MeanDiffusionKurtosisModel(gtab)
MSDKI can then be fitted to the synthetic data by calling the fit function
of this object:
msdki_fit = msdki_model.fit(dwi)
From the above fit object we can extract the two main parameters of the MSDKI, i.e.: 1) the mean signal diffusion (MSD); and 2) the mean signal kurtosis (MSK)
MSD = msdki_fit.msd
MSK = msdki_fit.msk
For a reference, we also calculate the mean diffusivity (MD) and mean
kurtosis (MK) from the standard DKI.
dki_model = dki.DiffusionKurtosisModel(gtab)
dki_fit = dki_model.fit(dwi)
MD = dki_fit.md
MK = dki_fit.mk(0, 3)
Now we plot the results as a function of the ground truth intersection angle and for different volume fractions of intra-cellular water.
fig1, axs = plt.subplots(nrows=2, ncols=2, figsize=(10, 10))
for f_i in range(f.size):
axs[0, 0].plot(ang, MSD[f_i], linewidth=1.0,
label=':math:`F: %.2f`' % f[f_i])
axs[0, 1].plot(ang, MSK[f_i], linewidth=1.0,
label=':math:`F: %.2f`' % f[f_i])
axs[1, 0].plot(ang, MD[f_i], linewidth=1.0,
label=':math:`F: %.2f`' % f[f_i])
axs[1, 1].plot(ang, MK[f_i], linewidth=1.0,
label=':math:`F: %.2f`' % f[f_i])
# Adjust properties of the first panel of the figure
axs[0, 0].set_xlabel('Intersection angle')
axs[0, 0].set_ylabel('MSD')
axs[0, 1].set_xlabel('Intersection angle')
axs[0, 1].set_ylabel('MSK')
axs[0, 1].legend(loc='center left', bbox_to_anchor=(1, 0.5))
axs[1, 0].set_xlabel('Intersection angle')
axs[1, 0].set_ylabel('MD')
axs[1, 1].set_xlabel('Intersection angle')
axs[1, 1].set_ylabel('MK')
axs[1, 1].legend(loc='center left', bbox_to_anchor=(1, 0.5))
plt.show()
fig1.savefig('MSDKI_simulations.png')
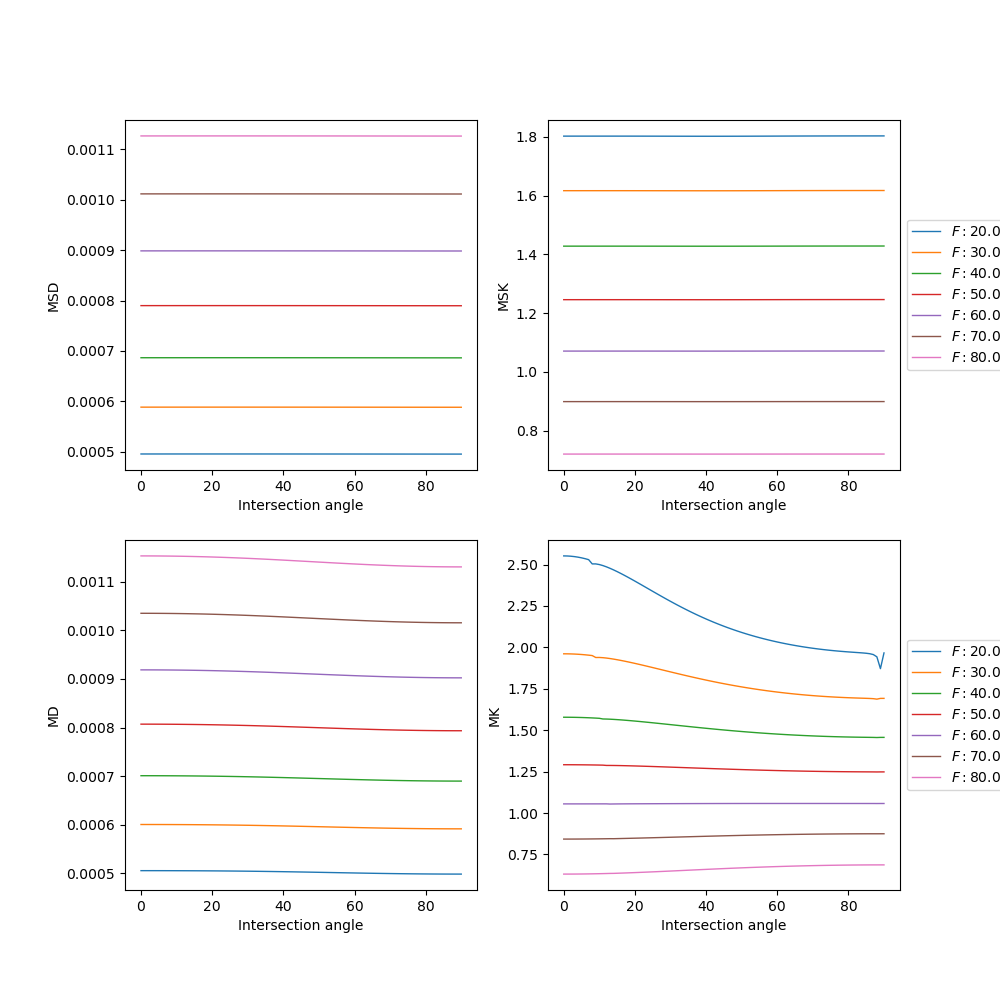
MSDKI and DKI measures for data of two crossing synthetic fibers. Upper panels show the MSDKI measures: 1) mean signal diffusivity (left panel); and 2) mean signal kurtosis (right panel). For reference, lower panels show the measures obtained by standard DKI: 1) mean diffusivity (left panel); and 2) mean kurtosis (right panel). All estimates are plotted as a function of the intersecting angle of the two crossing fibers. Different curves correspond to different ground truth axonal volume fraction of intra-cellular space.¶
The results of the above figure, demonstrate that both MSD and MSK are sensitive to axonal volume fraction (i.e. a microstructure property) but are independent to the intersectiong angle of the two crossing fibers (i.e. independent to properties regarding fiber orientation). In contrast, DKI measures seem to be independent to both axonal volume fraction and intersection angle.
Reconstructing diffusion data using MSDKI¶
Now that the properties of MSDKI were illustrated, let’s apply MSDKI to in-vivo diffusion-weighted data. As the example for the standard DKI (see Reconstruction of the diffusion signal with the kurtosis tensor model), we use fetch to download a multi-shell dataset which was kindly provided by Hansen and Jespersen (more details about the data are provided in their paper [Hansen2016]). The total size of the downloaded data is 192 MBytes, however you only need to fetch it once.
fetch_cfin_multib()
img, gtab = read_cfin_dwi()
data = img.get_data()
affine = img.affine
Before fitting the data, we preform some data pre-processing. For illustration, we only mask the data to avoid unnecessary calculations on the background of the image; however, you could also apply other pre-processing techniques. For example, some state of the art denoising algorithms are available in DIPY (e.g. the non-local means filter example-denoise-nlmeans or the local pca example-denoise-localpca).
maskdata, mask = median_otsu(data, vol_idx=[0, 1], median_radius=4, numpass=2,
autocrop=False, dilate=1)
Now that we have loaded and pre-processed the data we can go forward with MSDKI fitting. As for the synthetic data, the MSDKI model has to be first defined for the data’s GradientTable object:
msdki_model = msdki.MeanDiffusionKurtosisModel(gtab)
The data can then be fitted by calling the fit function of this object:
msdki_fit = msdki_model.fit(data, mask=mask)
Let’s then extract the two main MSDKI’s parameters: 1) mean signal diffusion (MSD); and 2) mean signal kurtosis (MSK).
MSD = msdki_fit.msd
MSK = msdki_fit.msk
For comparison, we calculate also the mean diffusivity (MD) and mean kurtosis (MK) from the standard DKI.
dki_model = dki.DiffusionKurtosisModel(gtab)
dki_fit = dki_model.fit(data, mask=mask)
MD = dki_fit.md
MK = dki_fit.mk(0, 3)
Let’s now visualize the data using matplotlib for a selected axial slice.
axial_slice = 9
fig2, ax = plt.subplots(2, 2, figsize=(6, 6),
subplot_kw={'xticks': [], 'yticks': []})
fig2.subplots_adjust(hspace=0.3, wspace=0.05)
ax.flat[0].imshow(MSD[:, :, axial_slice].T, cmap='gray', vmin=0, vmax=2.0e-3,
origin='lower')
ax.flat[0].set_title('MSD (MSDKI)')
ax.flat[1].imshow(MSK[:, :, axial_slice].T, cmap='gray', vmin=0, vmax=2,
origin='lower')
ax.flat[1].set_title('MSK (MSDKI)')
ax.flat[2].imshow(MD[:, :, axial_slice].T, cmap='gray', vmin=0, vmax=2.0e-3,
origin='lower')
ax.flat[2].set_title('MD (DKI)')
ax.flat[3].imshow(MK[:, :, axial_slice].T, cmap='gray', vmin=0, vmax=2,
origin='lower')
ax.flat[3].set_title('MK (DKI)')
plt.show()
fig2.savefig('MSDKI_invivo.png')
This figure shows that the contrast of in-vivo MSD and MSK maps (upper panels) are similar to the contrast of MD and MSK maps (lower panels); however, in the upper part we insure that direct contributions of fiber dispersion were removed. The upper panels also reveal that MSDKI measures are let sensitive to noise artefacts than standard DKI measures (as pointed by [NetoHe2018]), particularly one can appriciate that MSK maps always present positive values in brain white matter regions, while implausible negative kurtosis values are present in the MK maps in the same regions.
References¶
- NetoHe2019
Neto Henriques R, Jespersen SN, Shemesh N (2019). Microscopic anisotropy misestimation in spherical‐mean single diffusion encoding MRI. Magnetic Resonance in Medicine (In press). doi: 10.1002/mrm.27606
- Price2017
Price D, Tyler LK, Neto Henriques R, Campbell KR, Williams N, Treder M, Taylor J, Cam-CAN, Henson R (2017). Age-Related Delay in Visual and Auditory Evoked Responses is Mediated by White- and Gray-matter Differences. Nature Communications 8, 15671. doi: 10.1038/ncomms15671.
- Jensen2005
Jensen JH, Helpern JA, Ramani A, Lu H, Kaczynski K (2005). Diffusional Kurtosis Imaging: The Quantification of Non_Gaussian Water Diffusion by Means of Magnetic Resonance Imaging. Magnetic Resonance in Medicine 53: 1432-1440
- NetoHe2015(1,2)
Neto Henriques R, Correia MM, Nunes RG, Ferreira HA (2015). Exploring the 3D geometry of the diffusion kurtosis tensor - Impact on the development of robust tractography procedures and novel biomarkers, NeuroImage 111: 85-99
- NetoHe2018(1,2,3)
Henriques RN, 2018. Advanced Methods for Diffusion MRI Data Analysis and their Application to the Healthy Ageing Brain (Doctoral thesis). Downing College, University of Cambridge. https://doi.org/10.17863/CAM.29356
- Hansen2016
Hansen, B, Jespersen, SN (2016). Data for evaluation of fast kurtosis strategies, b-value optimization and exploration of diffusion MRI contrast. Scientific Data 3: 160072 doi:10.1038/sdata.2016.72
Example source code
You can download the full source code of this example. This same script is also included in the dipy source distribution under the doc/examples/ directory.