Note
Go to the end to download the full example code
DKI MultiTensor Simulation#
In this example we show how to simulate the Diffusion Kurtosis Imaging (DKI) data of a single voxel. DKI captures information about the non-Gaussian properties of water diffusion which is a consequence of the existence of tissue barriers and compartments. In these simulations compartmental heterogeneity is taken into account by modeling different compartments for the intra- and extra-cellular media of two populations of fibers. These simulations are performed according to [1].
We first import all relevant modules.
import matplotlib.pyplot as plt
import numpy as np
from dipy.core.gradients import gradient_table
from dipy.data import get_fnames
from dipy.io.gradients import read_bvals_bvecs
from dipy.reconst.dti import decompose_tensor, from_lower_triangular
from dipy.sims.voxel import multi_tensor_dki, single_tensor
For the simulation we will need a GradientTable with the b-values and
b-vectors. Here we use the GradientTable of the sample DIPY dataset
small_64D.
DKI requires data from more than one non-zero b-value. Since the dataset
small_64D was acquired with one non-zero b-value we artificially produce
a second non-zero b-value.
bvals = np.concatenate((bvals, bvals * 2), axis=0)
bvecs = np.concatenate((bvecs, bvecs), axis=0)
The b-values and gradient directions are then converted to DIPY’s
GradientTable format.
gtab = gradient_table(bvals, bvecs=bvecs)
In mevals we save the eigenvalues of each tensor. To simulate crossing
fibers with two different media (representing intra and extra-cellular
media), a total of four components have to be taken in to account (i.e. the
first two compartments correspond to the intra and extra cellular media for
the first fiber population while the others correspond to the media of the
second fiber population)
mevals = np.array(
[
[0.00099, 0, 0],
[0.00226, 0.00087, 0.00087],
[0.00099, 0, 0],
[0.00226, 0.00087, 0.00087],
]
)
In angles we save in polar coordinates (\(\theta, \phi\)) the
principal axis of each compartment tensor. To simulate crossing fibers at
70:math:^{circ} the compartments of the first fiber are aligned to the X-axis
while the compartments of the second fiber are aligned to the X-Z plane with
an angular deviation of 70:math:^{\circ} from the first one.
angles = [(90, 0), (90, 0), (20, 0), (20, 0)]
In fractions we save the percentage of the contribution of each
compartment, which is computed by multiplying the percentage of contribution
of each fiber population and the water fraction of each different medium
Having defined the parameters for all tissue compartments, the elements of
the diffusion tensor (DT), the elements of the kurtosis tensor (KT) and the
DW signals simulated from the DKI model can be obtain using the function
multi_tensor_dki.
We can also add Rician noise with a specific SNR.
For comparison purposes, we also compute the DW signal if only the diffusion
tensor components are taken into account. For this we use DIPY’s function
single_tensor which requires that dt is decomposed into its eigenvalues
and eigenvectors.
dt_evals, dt_evecs = decompose_tensor(from_lower_triangular(dt))
signal_dti = single_tensor(gtab, S0=200, evals=dt_evals, evecs=dt_evecs, snr=None)
Finally, we can visualize the values of the different version of simulated signals for all assumed gradient directions and bvalues.
plt.plot(signal_dti, label="noiseless dti")
plt.plot(signal_dki, label="noiseless dki")
plt.plot(signal_noisy, label="with noise")
plt.legend()
plt.savefig("simulated_dki_signal.png", bbox_inches="tight")
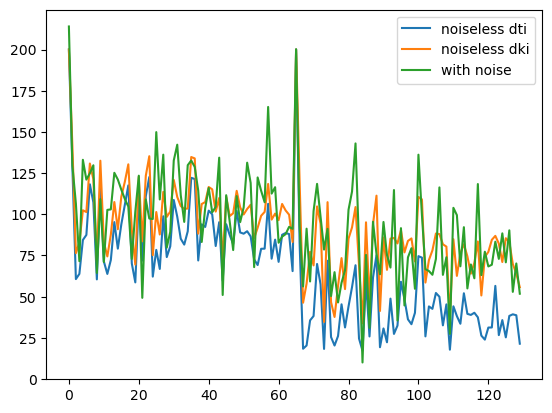
Simulated signals obtain from the DTI and DKI models.
Non-Gaussian diffusion properties in tissues are responsible to smaller signal attenuation for larger bvalues when compared to signal attenuation from free Gaussian water diffusion. This feature can be shown from the figure above, since signals simulated from the DKI models reveals larger DW signal intensities than the signals obtained only from the diffusion tensor components.
References#
Total running time of the script: (0 minutes 0.188 seconds)