Note
Go to the end to download the full example code
Extracting AFQ tract profiles from segmented bundles#
In this example, we will extract the values of a statistic from a volume, using the coordinates along the length of a bundle. These are called tract profiles
One of the challenges of extracting tract profiles is that some of the streamlines in a bundle may diverge significantly from the bundle in some locations. To overcome this challenge, we will use a strategy similar to that described in [1]: We will weight the contribution of each streamline to the bundle profile based on how far this streamline is from the mean trajectory of the bundle at that location.
import os.path as op
import matplotlib.pyplot as plt
import numpy as np
from dipy.data import fetch_hbn, get_two_hcp842_bundles
from dipy.io.image import load_nifti
from dipy.io.streamline import load_trk
from dipy.segment.clustering import QuickBundles
from dipy.segment.featurespeed import ResampleFeature
from dipy.segment.metricspeed import AveragePointwiseEuclideanMetric
import dipy.stats.analysis as dsa
import dipy.tracking.streamline as dts
To demonstrate this, we will use data from the Healthy Brain Network (HBN) study [2] that has already been processed [3]. For this demonstration, we will use only the left arcuate fasciculus (ARC) and the left corticospinal tract (CST) from the subject NDARAA948VFH.
We can use the dipy.io API to read in the bundles from file. load_trk returns both the streamlines, as well as header information, and the streamlines attribute will give us access to the sequence of arrays that contain the streamline coordinates.
cst_l_file = op.join(
afq_path,
"clean_bundles",
f"sub-{subject}_ses-{session}_acq-64dir_space-T1w_desc-preproc_dwi_space"
"-RASMM_model-CSD_desc-prob-afq-CST_L_tractography.trk",
)
arc_l_file = op.join(
afq_path,
"clean_bundles",
f"sub-{subject}_ses-{session}_acq-64dir_space-T1w_desc-preproc_dwi_space"
"-RASMM_model-CSD_desc-prob-afq-ARC_L_tractography.trk",
)
cst_l = load_trk(cst_l_file, reference="same", bbox_valid_check=False).streamlines
arc_l = load_trk(arc_l_file, reference="same", bbox_valid_check=False).streamlines
In the next step, we need to make sure that all the streamlines in each bundle are oriented the same way. For example, for the CST, we want to make sure that all the bundles have their cortical termination at one end of the streamline. This is so that when we later extract values from a volume, we will not have different streamlines going in opposite directions.
To orient all the streamlines in each bundle, we will create standard streamlines, by finding the centroids of the left ARC and CST bundle models.
The advantage of using the model bundles is that we can use the same standard for different subjects, which means that we’ll get the same orientation of the streamlines in all subjects.
model_arc_l_file, model_cst_l_file = get_two_hcp842_bundles()
model_arc_l = load_trk(
model_arc_l_file, reference="same", bbox_valid_check=False
).streamlines
model_cst_l = load_trk(
model_cst_l_file, reference="same", bbox_valid_check=False
).streamlines
feature = ResampleFeature(nb_points=100)
metric = AveragePointwiseEuclideanMetric(feature)
Since we are going to include all of the streamlines in the single cluster from the streamlines, we set the threshold to np.inf. We pull out the centroid as the standard using QuickBundles [4].
qb = QuickBundles(threshold=np.inf, metric=metric)
cluster_cst_l = qb.cluster(model_cst_l)
standard_cst_l = cluster_cst_l.centroids[0]
cluster_af_l = qb.cluster(model_arc_l)
standard_af_l = cluster_af_l.centroids[0]
We use the centroid streamline for each atlas bundle as the standard to orient all of the streamlines in each bundle from the individual subject. Here, the affine used is the one from the transform between the atlas and individual tractogram. This is so that the orienting is done relative to the space of the individual, and not relative to the atlas space.
oriented_cst_l = dts.orient_by_streamline(cst_l, standard_cst_l)
oriented_arc_l = dts.orient_by_streamline(arc_l, standard_af_l)
Tract profiles are created from a scalar property of the volume. Here, we read volumetric data from an image corresponding to the FA calculated in this subject with the diffusion tensor imaging (DTI) model.
fa, fa_affine = load_nifti(
op.join(
afq_path,
f"sub-{subject}_ses-{session}_acq-64dir_space-T1w_desc"
"-preproc_dwi_model-DTI_FA.nii.gz",
)
)
As mentioned at the outset, we would like to downweight the streamlines that are far from the core trajectory of the tracts. We calculate weights for each bundle:
w_cst_l = dsa.gaussian_weights(oriented_cst_l)
w_arc_l = dsa.gaussian_weights(oriented_arc_l)
And then use the weights to calculate the tract profiles for each bundle
profile_cst_l = dsa.afq_profile(fa, oriented_cst_l, affine=fa_affine, weights=w_cst_l)
profile_af_l = dsa.afq_profile(fa, oriented_arc_l, affine=fa_affine, weights=w_arc_l)
fig, (ax1, ax2) = plt.subplots(1, 2)
ax1.plot(profile_cst_l)
ax1.set_ylabel("Fractional anisotropy")
ax1.set_xlabel("Node along CST")
ax2.plot(profile_af_l)
ax2.set_xlabel("Node along ARC")
fig.savefig("tract_profiles")
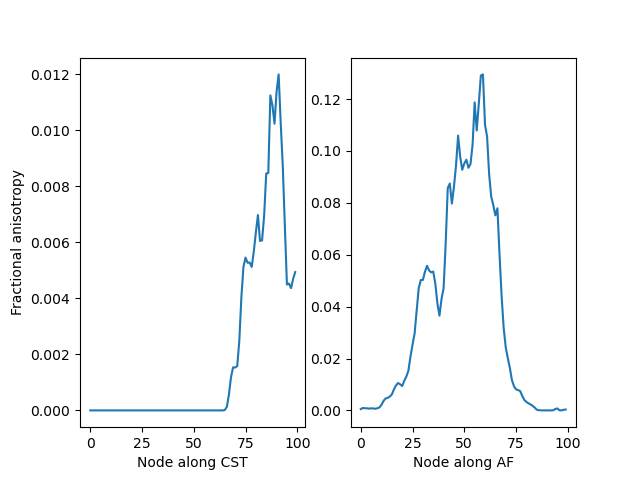
Bundle profiles for the fractional anisotropy in the left CST (left) and left AF (right).
References#
Total running time of the script: (0 minutes 16.812 seconds)