Note
Go to the end to download the full example code
Reconstruction with Multi-Shell Multi-Tissue CSD#
This example shows how to use Multi-Shell Multi-Tissue Constrained Spherical Deconvolution (MSMT-CSD) introduced by Jeurissen et al.[1]. This tutorial goes through the steps involved in implementing the method.
This method provides improved White Matter(WM), Grey Matter (GM), and Cerebrospinal fluid (CSF) volume fraction maps, which is otherwise overestimated in the standard CSD (SSST-CSD). This is done by using b-value dependencies of the different tissue types to estimate ODFs. This method thus extends the SSST-CSD introduced in [2].
The reconstruction of the fiber orientation distribution function (fODF) in MSMT-CSD involves the following steps:
Generate a mask using Median Otsu (optional step)
Denoise the data using MP-PCA (optional step)
Generate Anisotropic Powermap (if T1 unavailable)
Fit DTI model to the data
Tissue Classification (needs to be at least two classes of tissues)
Estimation of the fiber response function
Use the response function to reconstruct the fODF
First, we import all the modules we need from dipy as follows:
import matplotlib.pyplot as plt
import numpy as np
from dipy.core.gradients import gradient_table, unique_bvals_tolerance
from dipy.data import get_fnames, get_sphere
from dipy.denoise.localpca import mppca
import dipy.direction.peaks as dp
from dipy.io.gradients import read_bvals_bvecs
from dipy.io.image import load_nifti
from dipy.reconst.mcsd import (
MultiShellDeconvModel,
auto_response_msmt,
mask_for_response_msmt,
multi_shell_fiber_response,
response_from_mask_msmt,
)
import dipy.reconst.shm as shm
from dipy.segment.mask import median_otsu
from dipy.segment.tissue import TissueClassifierHMRF
from dipy.viz import actor, window
sphere = get_sphere(name="symmetric724")
For this example, we use fetch to download a multi-shell dataset which was kindly provided by Hansen and Jespersen (more details about the data are provided in their paper [3]). The total size of the downloaded data is 192 MBytes, however you only need to fetch it once.
fraw, fbval, fbvec, t1_fname = get_fnames(name="cfin_multib")
data, affine = load_nifti(fraw)
bvals, bvecs = read_bvals_bvecs(fbval, fbvec)
gtab = gradient_table(bvals, bvecs=bvecs)
For the sake of simplicity, we only select two non-zero b-values for this example.
The gradient table is also selected to have the selected b-values (0, 1000 and 2000)
gtab = gradient_table(bvals[sel_b], bvecs=bvecs[sel_b])
We make use of the median_otsu method to generate the mask for the data
as follows:
b0_mask, mask = median_otsu(data, median_radius=2, numpass=1, vol_idx=[0, 1])
print(data.shape)
(96, 96, 19, 67)
As one can see from its shape, the selected data contains a total of 67
volumes of images corresponding to all the diffusion gradient directions
of the selected b-values and call the mppca as follows:
denoised_arr = mppca(data, mask=mask, patch_radius=2)
C:\Users\skoudoro\AppData\Local\Continuum\Anaconda3\envs\py310\lib\site-packages\dipy\denoise\localpca.py:377: RuntimeWarning: invalid value encountered in divide
denoised_arr = thetax / theta
Now we will use the denoised array (denoised_arr) obtained from mppca
in the rest of the steps in the tutorial.
As for the next step, we generate the anisotropic powermap introduced by Dell’Acqua et al.[4]. To do so, we make use of the Q-ball Model as follows:
qball_model = shm.QballModel(gtab, 8)
We generate the peaks from the qball_model as follows:
peaks = dp.peaks_from_model(
model=qball_model,
data=denoised_arr,
relative_peak_threshold=0.5,
min_separation_angle=25,
sphere=sphere,
mask=mask,
)
ap = shm.anisotropic_power(peaks.shm_coeff)
plt.matshow(np.rot90(ap[:, :, 10]), cmap=plt.cm.bone)
plt.savefig("anisotropic_power_map.png")
plt.close()
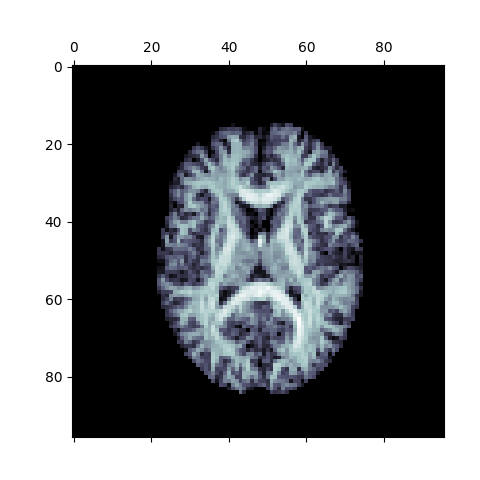
Anisotropic Power Map (Axial Slice)
print(ap.shape)
(96, 96, 19)
The above figure is a visualization of the axial slice of the Anisotropic Power Map. It can be treated as a pseudo-T1 for classification purposes using the Hidden Markov Random Fields (HMRF) classifier, if the T1 image is not available.
As we can see from the shape of the Anisotropic Power Map, it is 3D and can
be used for tissue classification using HMRF. The
HMRF needs the specification of the number of classes. For the case of
MSMT-CSD the nclass parameter needs to be >=2. In our case, we set
it to 3: namely corticospinal fluid (csf), white matter (wm) and gray
matter (gm).
nclass = 3
Then, the smoothness factor of the segmentation. Good performance is achieved with values between 0 and 0.5.
beta = 0.1
We then call the TissueClassifierHMRF with the parameters specified as
above:
hmrf = TissueClassifierHMRF()
initial_segmentation, final_segmentation, PVE = hmrf.classify(ap, nclass, beta)
>> Iteration: 0
>> Iteration: 1
>> Iteration: 2
>> Iteration: 3
>> Iteration: 4
>> Iteration: 5
>> Iteration: 6
>> Iteration: 7
>> Iteration: 8
>> Iteration: 9
>> Iteration: 10
>> Iteration: 11
>> Iteration: 12
>> Iteration: 13
>> Iteration: 14
>> Iteration: 15
>> Iteration: 16
>> Iteration: 17
>> Iteration: 18
>> Iteration: 19
>> Iteration: 20
>> Iteration: 21
>> Iteration: 22
>> Iteration: 23
Then, we get the tissues segmentation from the final_segmentation.
csf = np.where(final_segmentation == 1, 1, 0)
gm = np.where(final_segmentation == 2, 1, 0)
wm = np.where(final_segmentation == 3, 1, 0)
Now, we want the response function for each of the three tissues and for each
bvalues. This can be achieved in two different ways. If the case that tissue
segmentation is available or that one wants to see the tissue masks used to
compute the response functions, a combination of the functions
mask_for_response_msmt and response_from_mask is needed.
The mask_for_response_msmt function will return a mask of voxels within a
cuboid ROI and that meet some threshold constraints, for each tissue and
bvalue. More precisely, the WM mask must have a FA value above a given
threshold. The GM mask and CSF mask must have a FA below given thresholds
and a MD below other thresholds.
Note that for mask_for_response_msmt, the gtab and data should be for
bvalues under 1200, for optimal tensor fit.
mask_wm, mask_gm, mask_csf = mask_for_response_msmt(
gtab,
data,
roi_radii=10,
wm_fa_thr=0.7,
gm_fa_thr=0.3,
csf_fa_thr=0.15,
gm_md_thr=0.001,
csf_md_thr=0.0032,
)
C:\Users\skoudoro\AppData\Local\Continuum\Anaconda3\envs\py310\lib\site-packages\dipy\testing\decorators.py:192: UserWarning: Some b-values are higher than 1200.
The DTI fit might be affected.
return func(*args, **kwargs)
If one wants to use the previously computed tissue segmentation in addition to the threshold method, it is possible by simply multiplying both masks together.
mask_wm *= wm
mask_gm *= gm
mask_csf *= csf
The masks can also be used to calculate the number of voxels for each tissue.
nvoxels_wm = np.sum(mask_wm)
nvoxels_gm = np.sum(mask_gm)
nvoxels_csf = np.sum(mask_csf)
print(nvoxels_wm)
1407
Then, the response_from_mask function will return the msmt response
functions using precalculated tissue masks.
response_wm, response_gm, response_csf = response_from_mask_msmt(
gtab, data, mask_wm, mask_gm, mask_csf
)
Note that we can also get directly the response functions by calling the
auto_response_msmt function, which internally calls
mask_for_response_msmt followed by response_from_mask. By doing so,
we don’t have access to the masks and we might have problems with high
bvalues tensor fit.
auto_response_wm, auto_response_gm, auto_response_csf = auto_response_msmt(
gtab, data, roi_radii=10
)
C:\Users\skoudoro\AppData\Local\Continuum\Anaconda3\envs\py310\lib\site-packages\dipy\testing\decorators.py:192: UserWarning: Some b-values are higher than 1200.
The DTI fit might be affected. It is advised to use
mask_for_response_msmt with bvalues lower than 1200, followed by
response_from_mask_msmt with all bvalues to overcome this.
return func(*args, **kwargs)
As we can see below, adding the tissue segmentation can change the results of the response functions.
print("Responses")
print(response_wm)
print(response_gm)
print(response_csf)
print("Auto responses")
print(auto_response_wm)
print(auto_response_gm)
print(auto_response_csf)
Responses
[[1.56692320e-03 4.46085143e-04 4.46085143e-04 1.62131485e+02]
[1.34276461e-03 3.52534556e-04 3.52534556e-04 1.62131485e+02]]
[[9.72067588e-04 8.03258278e-04 8.03258278e-04 1.83944994e+02]
[8.84867552e-04 7.24112459e-04 7.24112459e-04 1.83944994e+02]]
[[1.48269864e-03 1.33687318e-03 1.33687318e-03 2.54087379e+02]
[1.19307328e-03 1.03487796e-03 1.03487796e-03 2.54087379e+02]]
Auto responses
[[1.70814058e-03 5.99497542e-04 5.99497542e-04 1.66589967e+02]
[1.37647490e-03 4.22844899e-04 4.22844899e-04 1.66589967e+02]]
[[1.02876747e-03 8.64284096e-04 8.64284096e-04 1.90155949e+02]
[9.05450074e-04 7.52760292e-04 7.52760292e-04 1.90155949e+02]]
[[1.33298612e-03 1.19461602e-03 1.19461602e-03 2.31346774e+02]
[1.09567957e-03 9.48548376e-04 9.48548376e-04 2.31346774e+02]]
At this point, there are two options on how to use those response functions.
We want to create a MultiShellDeconvModel, which takes a response function as
input. This response function can either be directly in the current format,
or it can be a MultiShellResponse format, as produced by the
multi_shell_fiber_response method. This function assumes a 3 compartments
model (wm, gm, csf) and takes one response function per tissue per bvalue. It
is important to note that the bvalues must be unique for this function.
ubvals = unique_bvals_tolerance(gtab.bvals)
response_mcsd = multi_shell_fiber_response(
sh_order_max=8,
bvals=ubvals,
wm_rf=response_wm,
gm_rf=response_gm,
csf_rf=response_csf,
)
As mentioned, we can also build the model directly and it will call
multi_shell_fiber_response internally. Important note here, the function
unique_bvals_tolerance is used to keep only unique bvalues from the gtab
given to the model, as input for multi_shell_fiber_response. This may
introduce differences between the calculated response of each method,
depending on the bvalues given to multi_shell_fiber_response externally.
response = np.array([response_wm, response_gm, response_csf])
mcsd_model_simple_response = MultiShellDeconvModel(gtab, response, sh_order_max=8)
Note that this technique only works for a 3 compartments model (wm, gm, csf).
If one wants more compartments, a custom MultiShellResponse object must be
used. It can be inspired by the multi_shell_fiber_response method.
Now we build the MSMT-CSD model with the response_mcsd as input. We then
call the fit function to fit one slice of the 3D data and visualize it.
mcsd_model = MultiShellDeconvModel(gtab, response_mcsd)
mcsd_fit = mcsd_model.fit(denoised_arr[:, :, 10:11])
C:\Users\skoudoro\AppData\Local\Continuum\Anaconda3\envs\py310\lib\site-packages\cvxpy\problems\problem.py:1403: UserWarning: Solution may be inaccurate. Try another solver, adjusting the solver settings, or solve with verbose=True for more information.
warnings.warn(
The volume fractions of tissues for each voxel are also accessible, as well
as the sh coefficients for all tissues. One can also get each sh tissue
separately using all_shm_coeff for each compartment (isotropic) and
shm_coeff for white matter.
The model allows one to predict a signal from sh coefficients. There are two ways of doing this.
mcsd_pred = mcsd_fit.predict()
mcsd_pred = mcsd_model.predict(mcsd_fit.all_shm_coeff)
From the fit obtained in the previous step, we generate the ODFs which can be visualized as follows:
mcsd_odf = mcsd_fit.odf(sphere)
print("ODF")
print(mcsd_odf.shape)
print(mcsd_odf[40, 40, 0])
fodf_spheres = actor.odf_slicer(
mcsd_odf, sphere=sphere, scale=1, norm=False, colormap="plasma"
)
interactive = False
scene = window.Scene()
scene.add(fodf_spheres)
scene.reset_camera_tight()
print("Saving illustration as msdodf.png")
window.record(scene=scene, out_path="msdodf.png", size=(600, 600))
if interactive:
window.show(scene)
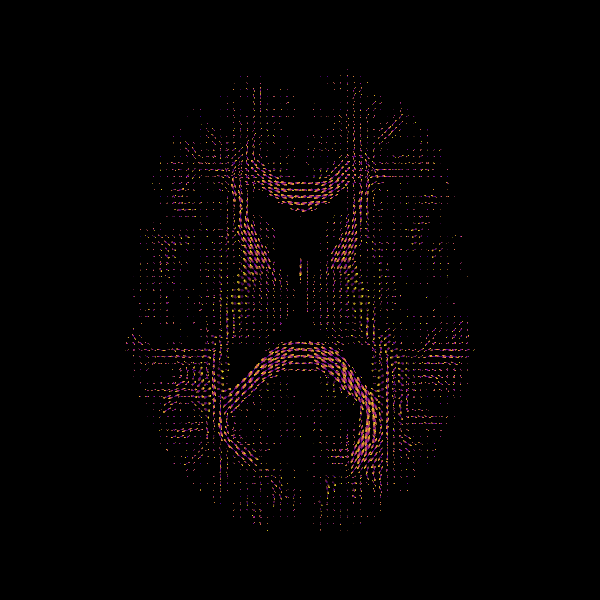
ODF
(96, 96, 1, 724)
[ 2.07819397e-03 2.48056889e-03 3.86317738e-03 5.80318112e-04
4.13450501e-03 1.90510861e-03 2.91056542e-04 2.78808950e-03
4.18908921e-05 3.61958970e-03 3.17237838e-03 1.23186090e-03
1.38676232e-03 8.99152866e-05 3.14832163e-04 1.06892957e-03
1.41825726e-03 3.65070379e-03 2.38250941e-03 9.38942356e-04
-4.67702568e-04 -2.34357229e-04 3.36801020e-03 1.26912924e-03
3.69950448e-03 -9.43861871e-04 7.76332334e-04 -2.76965057e-04
1.30105513e-03 1.04356019e-03 5.12702942e-03 2.57674067e-03
4.04510936e-03 2.23442424e-03 1.49056228e-04 1.99149700e-03
1.66042016e-04 3.10946414e-03 -4.39300078e-04 1.83495625e-03
1.67843779e-03 1.53254789e-02 6.33471409e-04 6.11565718e-03
8.00278023e-04 4.98227330e-03 4.41515801e-03 5.36097977e-04
1.15859621e-03 1.28331076e-02 9.97839734e-04 2.42637736e-03
2.32416120e-03 3.95430344e-03 3.73620758e-02 9.07359260e-04
4.61738361e-03 1.29678673e-03 2.62750988e-03 1.58723140e-03
5.94548779e-04 2.10784012e-03 5.20042914e-02 5.50135856e-04
5.07379628e-03 1.28699862e-03 3.87638912e-03 4.55356793e-02
5.39732048e-04 2.96030402e-03 2.09439424e-02 1.07074878e-04
-5.64342252e-04 8.94125547e-04 2.86817699e-03 1.04562301e-01
9.90321192e-04 4.46903789e-03 9.94198316e-04 1.67351437e-03
3.07922379e-02 -5.42989381e-05 3.02894434e-03 8.06017373e-02
3.00443786e-04 1.01026621e-03 9.62710991e-04 2.05185017e-03
1.29846537e-01 3.48944537e-04 2.69262470e-03 1.72415715e-02
-1.83042591e-05 1.02239573e-02 1.61051689e-04 2.71427461e-03
1.70664466e-01 1.20885237e-03 1.81182893e-03 3.72282936e-04
8.35450334e-04 1.03993364e-01 6.83646689e-04 2.88310692e-03
7.98688769e-02 1.12299493e-03 1.16502966e-03 4.87089565e-04
9.11524042e-04 2.34079283e-01 -1.65542623e-05 4.88922363e-04
8.12219624e-03 6.70655936e-04 5.23346100e-02 3.43813347e-03
3.15633085e-03 1.97143551e-01 2.90622199e-03 1.01889036e-03
7.93710007e-05 1.52279438e-05 2.17271730e-01 1.25254560e-03
5.87380649e-04 5.43598600e-02 2.60994960e-03 1.65058430e-02
4.63470182e-03 1.22384636e-03 3.12773778e-01 8.85653822e-04
8.14586308e-04 1.77664617e-03 1.21709957e-03 1.36277507e-01
1.00149315e-02 2.15089758e-03 1.70157210e-01 5.43197131e-03
7.28292374e-03 2.31501996e-03 -3.25902169e-04 3.38710836e-01
5.73876799e-04 -3.15229642e-04 2.47936185e-02 3.56338317e-03
5.83792975e-02 1.77440136e-02 1.96635855e-03 3.26243670e-01
3.85722562e-03 8.64078001e-03 1.06838050e-04 1.04555163e-03
2.53992561e-01 1.45864228e-02 -1.04756400e-05 1.09634648e-01
6.54845037e-03 2.48961940e-02 1.38681294e-02 4.19466814e-04
4.19583387e-01 2.42192035e-04 7.49746735e-03 6.49303974e-03
3.41843434e-03 1.33030323e-01 3.63123941e-02 1.10201510e-03
2.68332713e-01 6.72911840e-03 2.44350286e-02 4.39052455e-03
6.86860441e-04 3.73857661e-01 1.33382036e-02 3.02057132e-03
5.03885597e-02 5.29565370e-03 5.82095643e-02 4.01674891e-02
1.40213897e-03 4.21585971e-01 1.75680535e-03 2.81140382e-02
6.34829726e-04 2.39614203e-03 2.33580077e-01 4.94860840e-02
-6.65061333e-05 1.70904466e-01 6.38132479e-03 4.45167177e-02
2.18533114e-02 1.13296334e-03 4.50528233e-01 8.23909465e-03
2.07433362e-02 1.45461738e-02 2.95002361e-03 1.10818063e-01
7.53487264e-02 7.02700617e-04 3.42558305e-01 3.29635527e-03
5.63540384e-02 4.46209339e-03 1.24832761e-03 3.35102386e-01
4.88873403e-02 6.82259677e-03 8.01056189e-02 2.98621188e-03
6.64901754e-02 5.95809057e-02 2.06006364e-03 4.48670256e-01
3.63229030e-03 5.61980546e-02 1.67269383e-03 1.00770942e-03
1.80023822e-01 1.02478689e-01 -2.59898050e-04 2.19738552e-01
2.13595424e-03 8.04725397e-02 2.31177763e-02 1.03230831e-03
4.00851960e-01 3.63056657e-02 3.32697472e-02 2.46096185e-02
1.48486417e-04 9.25061276e-02 1.11447111e-01 1.54846471e-03
3.66207618e-01 8.69827448e-04 9.88722223e-02 2.62563824e-03
-2.45950496e-04 2.50953589e-01 1.06737025e-01 7.63297210e-03
1.05470615e-01 -1.54984383e-04 9.20058485e-02 6.53633515e-02
1.93584964e-03 4.00639040e-01 2.04792098e-02 8.22611069e-02
3.35319468e-03 3.34433859e-04 1.25263335e-01 1.55943878e-01
2.72215430e-05 2.38473404e-01 -5.31554116e-04 1.29384830e-01
1.74368496e-02 -7.71571848e-04 2.98499400e-01 8.77486229e-02
3.76454989e-02 3.38067351e-02 1.90919442e-03 9.26599617e-02
1.27656479e-01 2.24225059e-03 3.30608450e-01 8.83647236e-03
1.39530201e-01 6.23380689e-04 2.02865238e-03 1.60562810e-01
1.71004378e-01 5.55224326e-03 1.17127333e-01 2.57579584e-03
1.33600452e-01 5.51661163e-02 -1.63708142e-04 2.99784929e-01
5.82597409e-02 9.44015531e-02 5.19105865e-03 1.07878237e-02
9.10037403e-02 1.87524277e-01 8.63229514e-04 2.18749803e-01
5.56688954e-03 1.79054622e-01 9.10357390e-03 2.45220433e-03
1.85215698e-01 1.51322362e-01 3.20937098e-02 3.85036124e-02
1.66356111e-02 1.13196136e-01 1.16839829e-01 9.33405933e-04
2.49660975e-01 3.33779555e-02 1.62828367e-01 -2.43129847e-04
2.14112316e-02 9.27402686e-02 2.17155893e-01 2.94753072e-03
1.09286280e-01 1.61274058e-02 1.80868616e-01 3.55720392e-02
1.38598149e-03 1.84370835e-01 1.10953917e-01 8.72244551e-02
6.11560295e-03 4.28019034e-02 8.27587415e-02 1.83522763e-01
8.39934601e-04 1.66755842e-01 2.42426690e-02 2.12527850e-01
2.83709433e-03 2.57704045e-02 9.48342871e-02 2.04452619e-01
2.09923080e-02 3.60720306e-02 4.72655725e-02 1.45779054e-01
8.54721773e-02 4.73331175e-04 1.52938382e-01 7.17779632e-02
1.58762993e-01 -1.04042480e-04 7.07066651e-02 5.70571341e-02
2.25624177e-01 1.14091580e-03 8.35477744e-02 3.99773766e-02
2.17731612e-01 1.65009051e-02 2.12298936e-02 8.90775556e-02
1.60863347e-01 6.51733843e-02 5.28036399e-03 9.78310481e-02
9.35960381e-02 1.46777795e-01 2.07819397e-03 2.48056889e-03
3.86317738e-03 5.80318112e-04 4.13450501e-03 1.90510861e-03
2.91056542e-04 2.78808950e-03 4.18908921e-05 3.61958970e-03
3.17237838e-03 1.23186090e-03 1.38676232e-03 8.99152866e-05
3.14832163e-04 1.06892957e-03 1.41825726e-03 3.65070379e-03
2.38250941e-03 9.38942356e-04 -4.67702568e-04 -2.34357229e-04
3.36801020e-03 1.26912924e-03 3.69950448e-03 -9.43861871e-04
7.76332334e-04 -2.76965057e-04 1.30105513e-03 1.04356019e-03
5.12702942e-03 2.57674067e-03 4.04510936e-03 2.23442424e-03
1.49056228e-04 1.99149700e-03 1.66042016e-04 3.10946414e-03
-4.39300078e-04 1.83495625e-03 1.67843779e-03 1.53254789e-02
6.33471409e-04 6.11565718e-03 8.00278023e-04 4.98227330e-03
4.41515801e-03 5.36097977e-04 1.15859621e-03 1.28331076e-02
9.97839734e-04 2.42637736e-03 2.32416120e-03 3.95430344e-03
3.73620758e-02 9.07359260e-04 4.61738361e-03 1.29678673e-03
2.62750988e-03 1.58723140e-03 5.94548779e-04 2.10784012e-03
5.20042914e-02 5.50135856e-04 5.07379628e-03 1.28699862e-03
3.87638912e-03 4.55356793e-02 5.39732048e-04 2.96030402e-03
2.09439424e-02 1.07074878e-04 -5.64342252e-04 8.94125547e-04
2.86817699e-03 1.04562301e-01 9.90321192e-04 4.46903789e-03
9.94198316e-04 1.67351437e-03 3.07922379e-02 -5.42989381e-05
3.02894434e-03 8.06017373e-02 3.00443786e-04 1.01026621e-03
9.62710991e-04 2.05185017e-03 1.29846537e-01 3.48944537e-04
2.69262470e-03 1.72415715e-02 -1.83042591e-05 1.02239573e-02
1.61051689e-04 2.71427461e-03 1.70664466e-01 1.20885237e-03
1.81182893e-03 3.72282936e-04 8.35450334e-04 1.03993364e-01
6.83646689e-04 2.88310692e-03 7.98688769e-02 1.12299493e-03
1.16502966e-03 4.87089565e-04 9.11524042e-04 2.34079283e-01
-1.65542623e-05 4.88922363e-04 8.12219624e-03 6.70655936e-04
5.23346100e-02 3.43813347e-03 3.15633085e-03 1.97143551e-01
2.90622199e-03 1.01889036e-03 7.93710007e-05 1.52279438e-05
2.17271730e-01 1.25254560e-03 5.87380649e-04 5.43598600e-02
2.60994960e-03 1.65058430e-02 4.63470182e-03 1.22384636e-03
3.12773778e-01 8.85653822e-04 8.14586308e-04 1.77664617e-03
1.21709957e-03 1.36277507e-01 1.00149315e-02 2.15089758e-03
1.70157210e-01 5.43197131e-03 7.28292374e-03 2.31501996e-03
-3.25902169e-04 3.38710836e-01 5.73876799e-04 -3.15229642e-04
2.47936185e-02 3.56338317e-03 5.83792975e-02 1.77440136e-02
1.96635855e-03 3.26243670e-01 3.85722562e-03 8.64078001e-03
1.06838050e-04 1.04555163e-03 2.53992561e-01 1.45864228e-02
-1.04756400e-05 1.09634648e-01 6.54845037e-03 2.48961940e-02
1.38681294e-02 4.19466814e-04 4.19583387e-01 2.42192035e-04
7.49746735e-03 6.49303974e-03 3.41843434e-03 1.33030323e-01
3.63123941e-02 1.10201510e-03 2.68332713e-01 6.72911840e-03
2.44350286e-02 4.39052455e-03 6.86860441e-04 3.73857661e-01
1.33382036e-02 3.02057132e-03 5.03885597e-02 5.29565370e-03
5.82095643e-02 4.01674891e-02 1.40213897e-03 4.21585971e-01
1.75680535e-03 2.81140382e-02 6.34829726e-04 2.39614203e-03
2.33580077e-01 4.94860840e-02 -6.65061333e-05 1.70904466e-01
6.38132479e-03 4.45167177e-02 2.18533114e-02 1.13296334e-03
4.50528233e-01 8.23909465e-03 2.07433362e-02 1.45461738e-02
2.95002361e-03 1.10818063e-01 7.53487264e-02 7.02700617e-04
3.42558305e-01 3.29635527e-03 5.63540384e-02 4.46209339e-03
1.24832761e-03 3.35102386e-01 4.88873403e-02 6.82259677e-03
8.01056189e-02 2.98621188e-03 6.64901754e-02 5.95809057e-02
2.06006364e-03 4.48670256e-01 3.63229030e-03 5.61980546e-02
1.67269383e-03 1.00770942e-03 1.80023822e-01 1.02478689e-01
-2.59898050e-04 2.19738552e-01 2.13595424e-03 8.04725397e-02
2.31177763e-02 1.03230831e-03 4.00851960e-01 3.63056657e-02
3.32697472e-02 2.46096185e-02 1.48486417e-04 9.25061276e-02
1.11447111e-01 1.54846471e-03 3.66207618e-01 8.69827448e-04
9.88722223e-02 2.62563824e-03 -2.45950496e-04 2.50953589e-01
1.06737025e-01 7.63297210e-03 1.05470615e-01 -1.54984383e-04
9.20058485e-02 6.53633515e-02 1.93584964e-03 4.00639040e-01
2.04792098e-02 8.22611069e-02 3.35319468e-03 3.34433859e-04
1.25263335e-01 1.55943878e-01 2.72215430e-05 2.38473404e-01
-5.31554116e-04 1.29384830e-01 1.74368496e-02 -7.71571848e-04
2.98499400e-01 8.77486229e-02 3.76454989e-02 3.38067351e-02
1.90919442e-03 9.26599617e-02 1.27656479e-01 2.24225059e-03
3.30608450e-01 8.83647236e-03 1.39530201e-01 6.23380689e-04
2.02865238e-03 1.60562810e-01 1.71004378e-01 5.55224326e-03
1.17127333e-01 2.57579584e-03 1.33600452e-01 5.51661163e-02
-1.63708142e-04 2.99784929e-01 5.82597409e-02 9.44015531e-02
5.19105865e-03 1.07878237e-02 9.10037403e-02 1.87524277e-01
8.63229514e-04 2.18749803e-01 5.56688954e-03 1.79054622e-01
9.10357390e-03 2.45220433e-03 1.85215698e-01 1.51322362e-01
3.20937098e-02 3.85036124e-02 1.66356111e-02 1.13196136e-01
1.16839829e-01 9.33405933e-04 2.49660975e-01 3.33779555e-02
1.62828367e-01 -2.43129847e-04 2.14112316e-02 9.27402686e-02
2.17155893e-01 2.94753072e-03 1.09286280e-01 1.61274058e-02
1.80868616e-01 3.55720392e-02 1.38598149e-03 1.84370835e-01
1.10953917e-01 8.72244551e-02 6.11560295e-03 4.28019034e-02
8.27587415e-02 1.83522763e-01 8.39934601e-04 1.66755842e-01
2.42426690e-02 2.12527850e-01 2.83709433e-03 2.57704045e-02
9.48342871e-02 2.04452619e-01 2.09923080e-02 3.60720306e-02
4.72655725e-02 1.45779054e-01 8.54721773e-02 4.73331175e-04
1.52938382e-01 7.17779632e-02 1.58762993e-01 -1.04042480e-04
7.07066651e-02 5.70571341e-02 2.25624177e-01 1.14091580e-03
8.35477744e-02 3.99773766e-02 2.17731612e-01 1.65009051e-02
2.12298936e-02 8.90775556e-02 1.60863347e-01 6.51733843e-02
5.28036399e-03 9.78310481e-02 9.35960381e-02 1.46777795e-01]
Saving illustration as msdodf.png
MSMT-CSD Peaks and ODFs.
References#
Total running time of the script: (6 minutes 4.564 seconds)