Note
Go to the end to download the full example code
Continuous and analytical diffusion signal modelling with 3D-SHORE#
We show how to model the diffusion signal as a linear combination of continuous functions from the SHORE basis [1], [2], [3]. We also compute the analytical Orientation Distribution Function (ODF).
First import the necessary modules:
from dipy.core.gradients import gradient_table
from dipy.data import get_fnames, get_sphere
from dipy.io.gradients import read_bvals_bvecs
from dipy.io.image import load_nifti
from dipy.reconst.shore import ShoreModel
from dipy.viz import actor, window
Download and read the data for this tutorial.
# ``fetch_isbi2013_2shell()`` provides data from the `ISBI HARDI contest 2013
# <http://hardi.epfl.ch/static/events/2013_ISBI/>`_ acquired for two shells at
# b-values 1500 :math:`s/mm^2` and 2500 :math:`s/mm^2`.
# The six parameters of these two functions define the ROI where to reconstruct
# the data. They respectively correspond to ``(xmin,xmax,ymin,ymax,zmin,zmax)``
# with x, y, z and the three axis defining the spatial positions of the voxels.
fraw, fbval, fbvec = get_fnames(name="isbi2013_2shell")
data, affine = load_nifti(fraw)
bvals, bvecs = read_bvals_bvecs(fbval, fbvec)
gtab = gradient_table(bvals, bvecs=bvecs)
data_small = data[10:40, 22, 10:40]
print(f"data.shape {data.shape}")
data.shape (50, 50, 50, 64)
data contains the voxel data and gtab contains a GradientTable
object (gradient information e.g. b-values). For example, to show the
b-values it is possible to write:
print(gtab.bvals)
Instantiate the SHORE Model.
radial_order is the radial order of the SHORE basis.
zeta is the scale factor of the SHORE basis.
lambdaN and lambdaL are the radial and angular regularization
constants, respectively.
For details regarding these four parameters see [4] and [1].
radial_order = 6
zeta = 700
lambdaN = 1e-8
lambdaL = 1e-8
asm = ShoreModel(
gtab, radial_order=radial_order, zeta=zeta, lambdaN=lambdaN, lambdaL=lambdaL
)
Fit the SHORE model to the data
Load an odf reconstruction sphere
sphere = get_sphere(name="repulsion724")
Compute the ODFs
odf = asmfit.odf(sphere)
print(f"odf.shape {odf.shape}")
odf.shape (30, 30, 724)
Display the ODFs
# Enables/disables interactive visualization
interactive = False
scene = window.Scene()
sfu = actor.odf_slicer(odf[:, None, :], sphere=sphere, colormap="plasma", scale=0.5)
sfu.RotateX(-90)
sfu.display(y=0)
scene.add(sfu)
window.record(scene=scene, out_path="odfs.png", size=(600, 600))
if interactive:
window.show(scene)
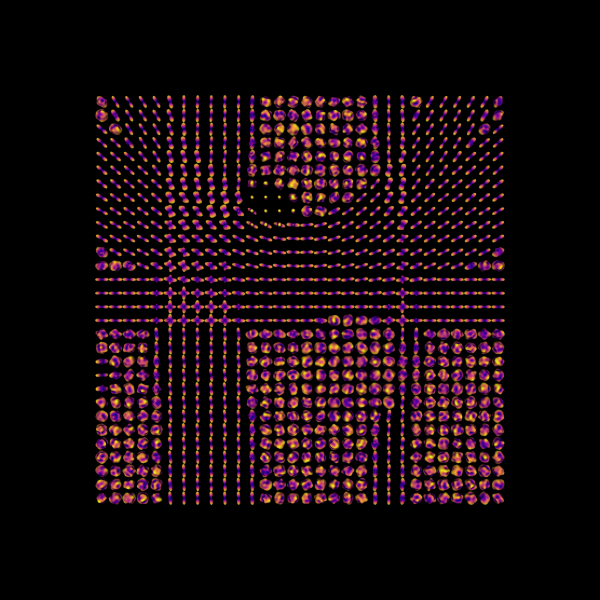
Orientation distribution functions.
References#
Total running time of the script: (0 minutes 2.452 seconds)