Note
Go to the end to download the full example code
Nonrigid Bundle Registration with BundleWarp#
This example explains how you can nonlinearly register two bundles from two different subjects directly in the space of streamlines [1], [2].
To show the concept, we will use two pre-saved uncinate fasciculus bundles. The algorithm used here is called BundleWarp, streamline-based nonlinear registration of white matter tracts [1].
from os.path import join as pjoin
from time import time
from dipy.align.streamwarp import (
bundlewarp,
bundlewarp_shape_analysis,
bundlewarp_vector_filed,
)
from dipy.data import fetch_bundle_warp_dataset
from dipy.io.stateful_tractogram import Space, StatefulTractogram
from dipy.io.streamline import load_trk, save_tractogram
from dipy.tracking.streamline import (
Streamlines,
set_number_of_points,
unlist_streamlines,
)
from dipy.viz.streamline import viz_displacement_mag, viz_two_bundles, viz_vector_field
Let’s download and load two uncinate fasciculus bundles in the left hemisphere of the brain (UF_L) available here: https://figshare.com/articles/dataset/Test_Bundles_for_DIPY/22557733
bundle_warp_files = fetch_bundle_warp_dataset()
s_UF_L_path = pjoin(bundle_warp_files[1], "s_UF_L.trk")
m_UF_L_path = pjoin(bundle_warp_files[1], "m_UF_L.trk")
uf_subj1 = load_trk(s_UF_L_path, reference="same", bbox_valid_check=False).streamlines
uf_subj2 = load_trk(m_UF_L_path, reference="same", bbox_valid_check=False).streamlines
Let’s resample the streamlines so that they both have the same number of points per streamline. Here we will use 20 points.
static = Streamlines(set_number_of_points(uf_subj1, nb_points=20))
moving = Streamlines(set_number_of_points(uf_subj2, nb_points=20))
We call uf_subj2 a moving bundle as it will be nonlinearly aligned with
uf_subj1 (static) bundle. Here is how this is done.
Let’s visualize static bundle in red and moving in green before registration.
viz_two_bundles(static, moving, fname="static_and_moving.png")

BundleWarp method provides a unique ability to either partially or fully deform a moving bundle by the use of a single regularization parameter alpha. alpha controls the trade-off between regularizing the deformation and having points match very closely. The lower the value of alpha, the more closely the bundles would match.
Let’s partially deform bundle by setting alpha=0.5.
time taken by BundleWarp registration in seconds = 5.768532037734985
Let’s visualize static bundle in red and moved (warped) in green. Note: You can set interactive=True in visualization functions throughout this tutorial if you prefer to get interactive visualization window.
viz_two_bundles(static, deformed_bundle, fname="static_and_partially_deformed.png")

Let’s visualize linearly moved bundle in blue and nonlinearly moved bundle in green to see BundleWarp registration improvement over linear SLR registration.
viz_two_bundles(
moving_aligned,
deformed_bundle,
fname="linearly_and_nonlinearly_moved.png",
c1=(0, 0, 1),
)

Now, let’s visualize deformation vector field generated by BundleWarp. This shows us visually where and how much and in what directions deformations were added by BundleWarp.
offsets, directions, colors = bundlewarp_vector_filed(moving_aligned, deformed_bundle)
points_aligned, _ = unlist_streamlines(moving_aligned)
Visualizing just the vector field.
fname = "partially_vectorfield.png"
viz_vector_field(points_aligned, directions, colors, offsets, fname)

Let’s visualize vector field over linearly moved bundle. This will show how much deformations were introduced after linear registration.
fname = "partially_vectorfield_over_linearly_moved.png"
viz_vector_field(
points_aligned, directions, colors, offsets, fname, bundle=moving_aligned
)

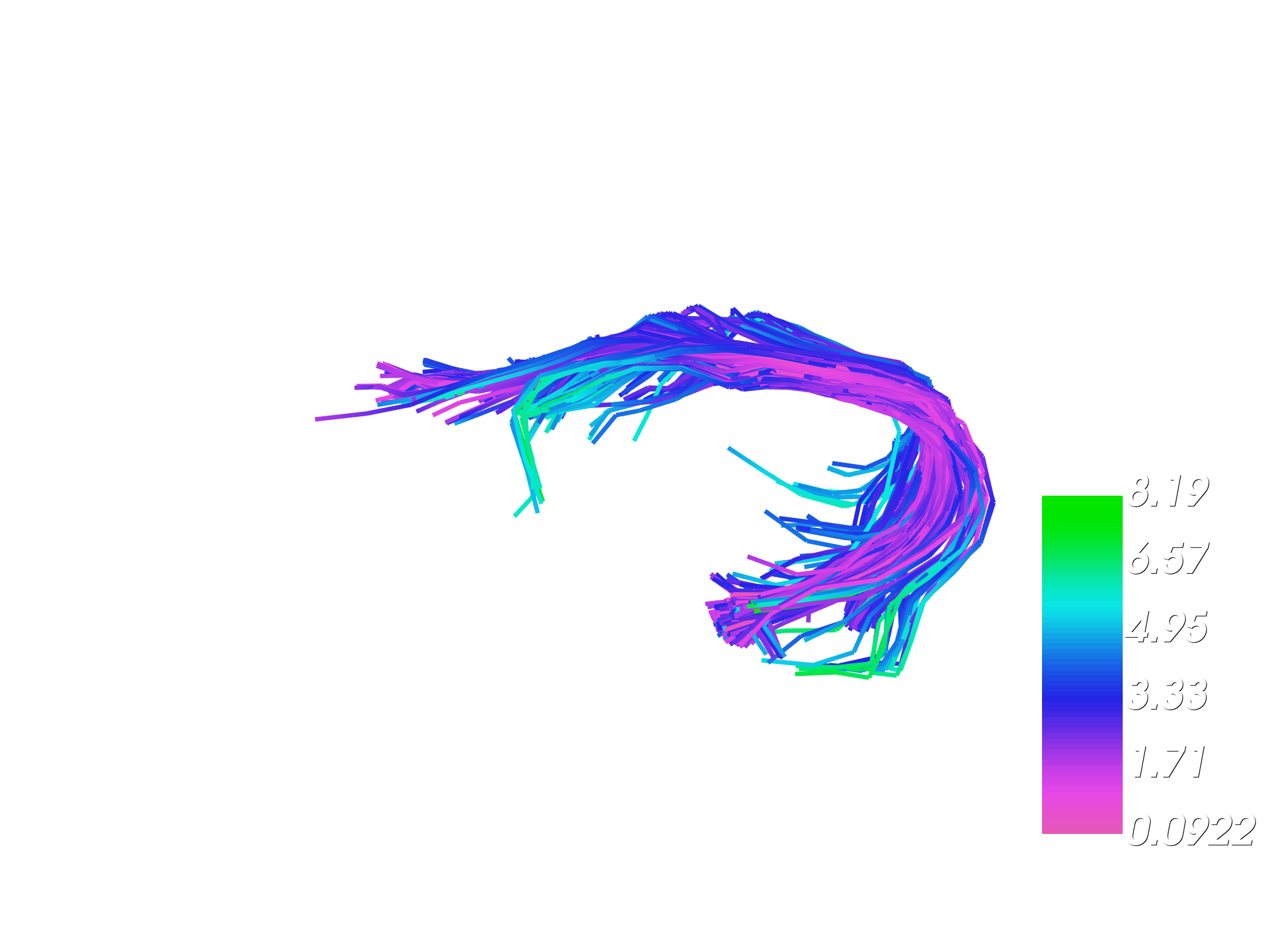
We can also visualize the magnitude of deformations in mm mapped over affinely moved bundle. It shows which streamlines were deformed the most after affine registration.
fname = "partially_deformation_magnitude_over_linearly_moved.png"
viz_displacement_mag(moving_aligned, offsets, fname, interactive=False)
Saving partially warped bundle.
new_tractogram = StatefulTractogram(deformed_bundle, m_UF_L_path, Space.RASMM)
save_tractogram(new_tractogram, "partially_deformed_bundle.trk", bbox_valid_check=False)
True
Let’s fully deform the moving bundle by setting alpha <= 0.01
We will use MDF distances computed and returned by previous run of BundleWarp method. This will save computation time.
C:\Users\skoudoro\AppData\Local\Continuum\Anaconda3\envs\py310\lib\site-packages\dipy\testing\decorators.py:192: UserWarning: Using alpha<=0.01 will result in extreme deformations
return func(*args, **kwargs)
using pre-computed distances
time taken by BundleWarp registration in seconds = 4.92172646522522
Let’s visualize static bundle in red and moved (completely warped) in green.
viz_two_bundles(static, deformed_bundle2, fname="static_and_fully_deformed.png")

Now, let’s visualize the deformation vector field generated by BundleWarp. This shows us visually where and how much and in what directions deformations were added by BundleWarp to perfectly warp moving bundle to look like static.
offsets, directions, colors = bundlewarp_vector_filed(moving_aligned, deformed_bundle2)
points_aligned, _ = unlist_streamlines(moving_aligned)
Visualizing just the vector field.
fname = "fully_vectorfield.png"
viz_vector_field(points_aligned, directions, colors, offsets, fname)

Let’s visualize vector field over linearly moved bundle. This will show how much deformations were introduced after linear registration by fully deforming the moving bundle.
fname = "fully_vectorfield_over_linearly_moved.png"
viz_vector_field(
points_aligned, directions, colors, offsets, fname, bundle=moving_aligned
)

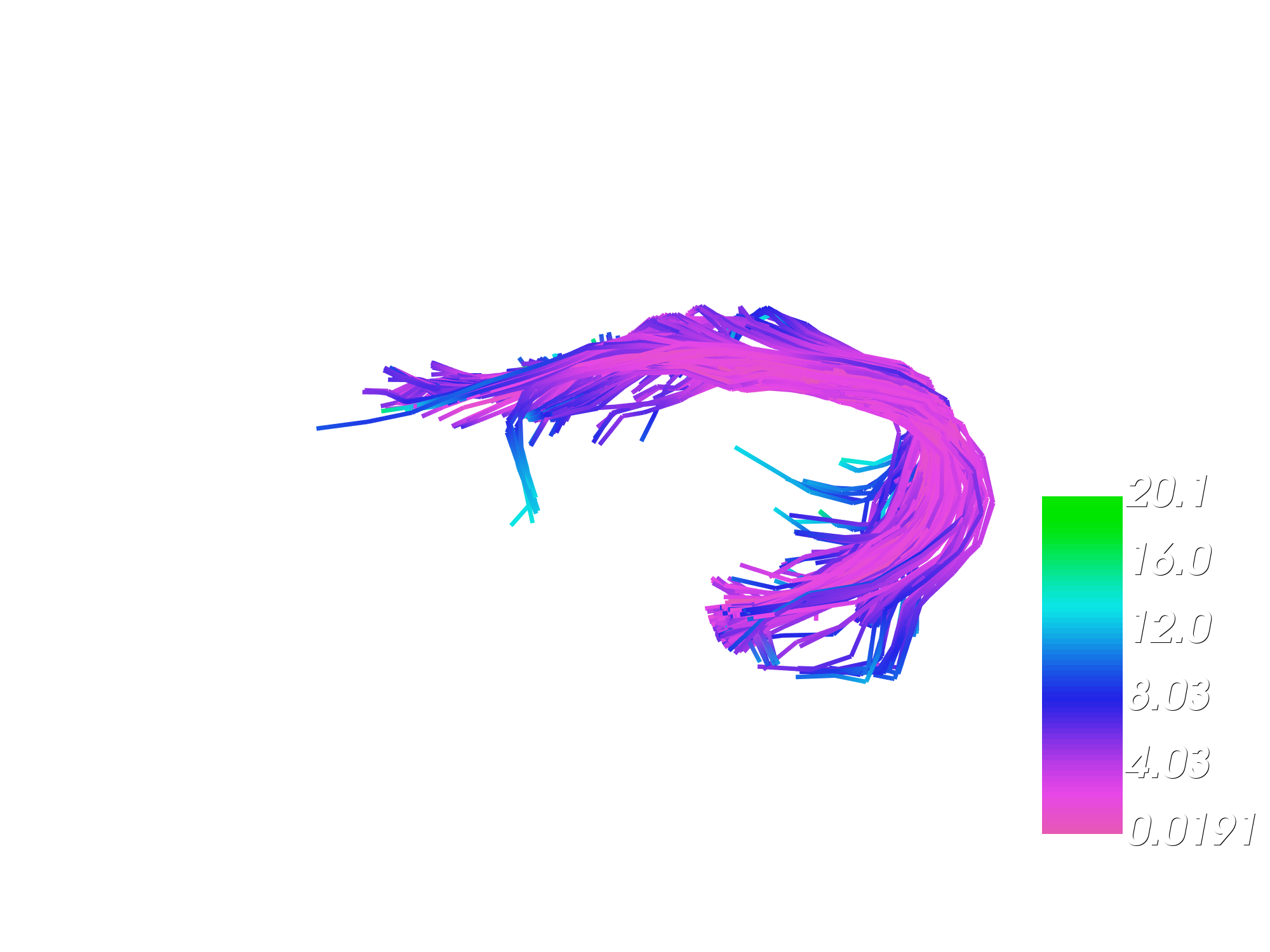
Let’s visualize the magnitude of deformations in mm mapped over affinely moved bundle. It shows which streamlines were deformed the most after affine registration.
fname = "fully_deformation_magnitude_over_linearly_moved.png"
viz_displacement_mag(moving_aligned, offsets, fname, interactive=False)
We can also perform bundle shape difference analysis using the displacement field generated by fully warping the moving bundle to look exactly like static bundle. Here, we plot bundle shape profile using BUAN. Bundle shape profile shows the average magnitude of deformations along the length of the bundle. Segments where we observe higher deformations are the areas where two bundles differ the most in shape.
_, _ = bundlewarp_shape_analysis(
moving_aligned, deformed_bundle, no_disks=10, plotting=False
)
Saving fully warped bundle.
new_tractogram = StatefulTractogram(deformed_bundle2, m_UF_L_path, Space.RASMM)
save_tractogram(new_tractogram, "fully_deformed_bundle.trk", bbox_valid_check=False)
True
References#
Total running time of the script: (0 minutes 17.863 seconds)