Note
Go to the end to download the full example code
SNR estimation for Diffusion-Weighted Images#
Computing the Signal-to-Noise-Ratio (SNR) of DW images is still an open question, as SNR depends on the white matter structure of interest as well as the gradient direction corresponding to each DWI.
In classical MRI, SNR can be defined as the ratio of the mean of the signal divided by the standard deviation of the underlying Gaussian noise, that is \(SNR = mean(signal) / std(noise)\). The noise standard deviation can be computed from the background in any of the DW images. How do we compute the mean of the signal, and what signal?
The strategy here is to compute a ‘worst-case’ SNR for DWI. Several white matter structures such as the corpus callosum (CC), corticospinal tract (CST), or the superior longitudinal fasciculus (SLF) can be easily identified from the colored-FA (CFA) map. In this example, we will use voxels from the CC, which have the characteristic of being highly red in the CFA map since they are mainly oriented in the left-right direction. We know that the DW image closest to the X-direction will be the one with the most attenuated diffusion signal. This is the strategy adopted in several recent papers (see [1] and [2]). It gives a good indication of the quality of the DWI data.
First, we compute the tensor model in a brain mask (see the Reconstruction of the diffusion signal with DTI (single tensor) model example for further explanations).
Let’s load the necessary modules:
import matplotlib.pyplot as plt
import numpy as np
from scipy.ndimage import binary_dilation
from dipy.core.gradients import gradient_table
from dipy.data import get_fnames
from dipy.io.gradients import read_bvals_bvecs
from dipy.io.image import load_nifti, save_nifti
from dipy.reconst.dti import TensorModel
from dipy.segment.mask import bounding_box, median_otsu, segment_from_cfa
Then, we fetch and load a specific dataset with 64 gradient directions:
hardi_fname, hardi_bval_fname, hardi_bvec_fname = get_fnames(name="stanford_hardi")
data, affine = load_nifti(hardi_fname)
bvals, bvecs = read_bvals_bvecs(hardi_bval_fname, hardi_bvec_fname)
gtab = gradient_table(bvals, bvecs=bvecs)
print("Computing brain mask...")
b0_mask, mask = median_otsu(data, vol_idx=[0])
print("Computing tensors...")
tenmodel = TensorModel(gtab)
tensorfit = tenmodel.fit(data, mask=mask)
Computing brain mask...
Computing tensors...
Next, we set our red-green-blue thresholds to (0.6, 1) in the x axis and (0, 0.1) in the y and z axes respectively. These values work well in practice to isolate the very RED voxels of the cfa map.
Then, as assurance, we want just RED voxels in the CC (there could be noisy red voxels around the brain mask and we don’t want those). Unless the brain acquisition was badly aligned, the CC is always close to the mid-sagittal slice.
The following lines perform these two operations and then saves the computed mask.
print("Computing worst-case/best-case SNR using the corpus callosum...")
threshold = (0.6, 1, 0, 0.1, 0, 0.1)
CC_box = np.zeros_like(data[..., 0])
mins, maxs = bounding_box(mask)
mins = np.array(mins)
maxs = np.array(maxs)
diff = (maxs - mins) // 4
bounds_min = mins + diff
bounds_max = maxs - diff
CC_box[
bounds_min[0] : bounds_max[0],
bounds_min[1] : bounds_max[1],
bounds_min[2] : bounds_max[2],
] = 1
mask_cc_part, cfa = segment_from_cfa(tensorfit, CC_box, threshold, return_cfa=True)
save_nifti("cfa_CC_part.nii.gz", (cfa * 255).astype(np.uint8), affine)
save_nifti("mask_CC_part.nii.gz", mask_cc_part.astype(np.uint8), affine)
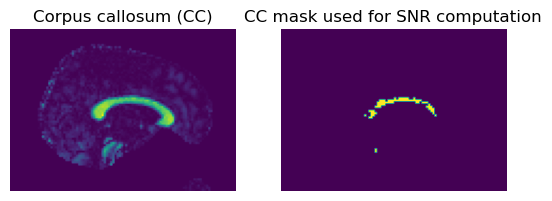
region = 40
fig = plt.figure("Corpus callosum segmentation")
plt.subplot(1, 2, 1)
plt.title("Corpus callosum (CC)")
plt.axis("off")
red = cfa[..., 0]
plt.imshow(np.rot90(red[region, ...]))
plt.subplot(1, 2, 2)
plt.title("CC mask used for SNR computation")
plt.axis("off")
plt.imshow(np.rot90(mask_cc_part[region, ...]))
fig.savefig("CC_segmentation.png", bbox_inches="tight")
Computing worst-case/best-case SNR using the corpus callosum...
Now that we are happy with our crude CC mask that selected voxels in the x-direction, we can use all the voxels to estimate the mean signal in this region.
mean_signal = np.mean(data[mask_cc_part], axis=0)
Now, we need a good background estimation. We will reuse the brain mask computed before and invert it to catch the outside of the brain. This could also be determined manually with a ROI in the background.
Warning
Certain MR manufacturers mask out the outside of the brain with 0’s. One thus has to be careful how the noise ROI is defined.
mask_noise = binary_dilation(mask, iterations=10)
mask_noise[..., : mask_noise.shape[-1] // 2] = 1
mask_noise = ~mask_noise
save_nifti("mask_noise.nii.gz", mask_noise.astype(np.uint8), affine)
noise_std = np.std(data[mask_noise, :])
print("Noise standard deviation sigma= ", noise_std)
Noise standard deviation sigma= 8.17113266785504
We can now compute the SNR for each DWI. For example, report SNR for DW images with gradient direction that lies the closest to the X, Y and Z axes.
# Exclude null bvecs from the search
idx = np.sum(gtab.bvecs, axis=-1) == 0
gtab.bvecs[idx] = np.inf
axis_X = np.argmin(np.sum((gtab.bvecs - np.array([1, 0, 0])) ** 2, axis=-1))
axis_Y = np.argmin(np.sum((gtab.bvecs - np.array([0, 1, 0])) ** 2, axis=-1))
axis_Z = np.argmin(np.sum((gtab.bvecs - np.array([0, 0, 1])) ** 2, axis=-1))
for direction in [0, axis_X, axis_Y, axis_Z]:
SNR = mean_signal[direction] / noise_std
if direction == 0:
print(f"SNR for the b=0 image is : {SNR}")
else:
print(f"SNR for direction {direction} {gtab.bvecs[direction]} is : {SNR}")
SNR for the b=0 image is : 47.366354266706736
SNR for direction 58 [ 0.98875 0.1177 -0.09229] is : 5.918432129721111
SNR for direction 57 [-0.05039 0.99871 0.0054406] is : 26.72068171809924
SNR for direction 126 [-0.11825 -0.039925 0.99218 ] is : 27.592653853373644
Since the CC is aligned with the X axis, the lowest SNR is for that gradient direction. In comparison, the DW images in the perpendicular Y and Z axes have a high SNR. The b0 still exhibits the highest SNR, since there is no signal attenuation.
Hence, we can say the Stanford diffusion data has a ‘worst-case’ SNR of approximately 5, a ‘best-case’ SNR of approximately 24, and a SNR of 42 on the b0 image.
References#
Total running time of the script: (0 minutes 18.687 seconds)